Fig. 1.
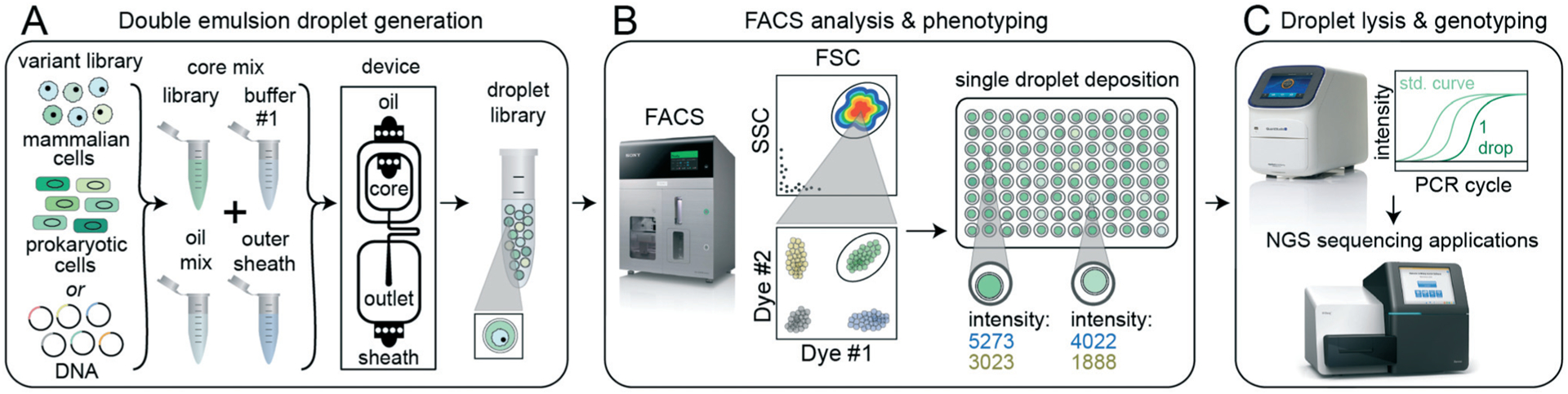
sdDE-FACS workflow. (A) Cell or DNA variants of interest are loaded into a DE droplet generator device to produce a library of droplets each containing a different variant. DEs can be generated for a wide variety of reactions by adjusting core mix reagents and buffers, number of core inlets, and droplet size. (B) DE droplets are analyzed via FACS to quantify morphology (FSC vs. SSC) and relevant fluorescence signals (by fluorescent intensity) and then sorted into wells of a multiwell plate. (C) Sorted DE droplets can be lysed to recover nucleic acids for downstream applications, such as qPCR or next-generation sequencing, that link droplet genotype to phenotype (e.g. enzymatic reaction turnover, presence of a specific cell type, or completion of a cellular reaction).
