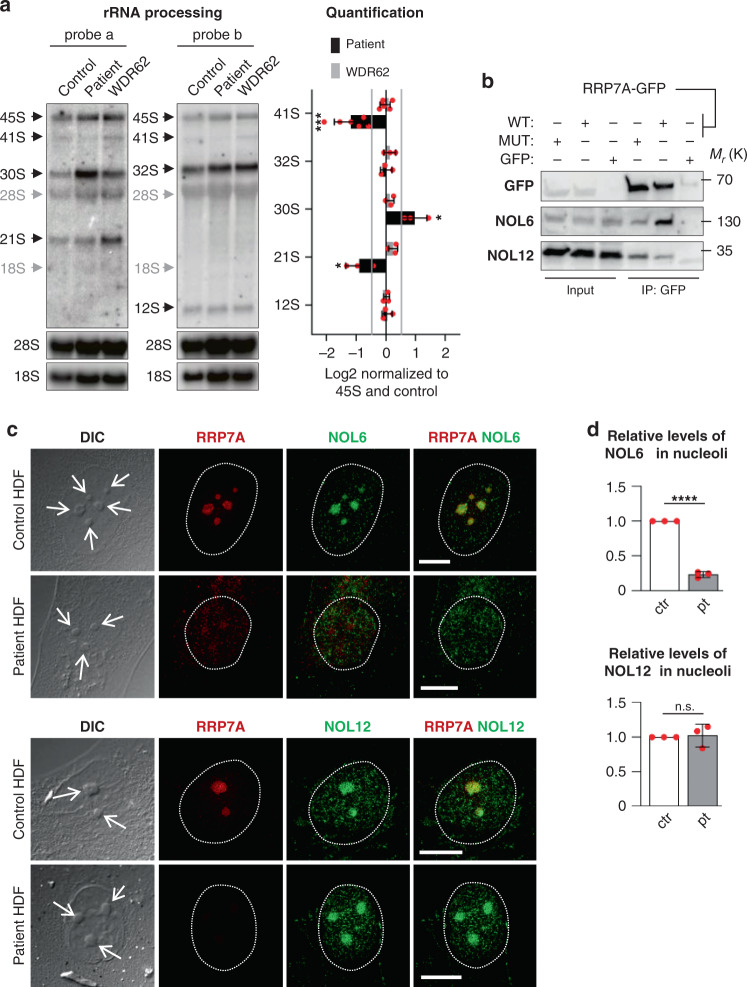
Fig. 5. Nucleolar localization and rRNA processing are disrupted in patient HDFs.
a Left: representative northern blots of parallel gel runs of total RNA samples from control and patient HDFs with mutations in RRP7A (patient) and WDR62 (MCPH2 pt.), respectively. Black arrows indicate processing intermediates inferred from the analyses and gray arrows mark the migration of mature rRNA species as inferred from reprobing of the filters with probes targeting these RNA species. a Right: quantification of northern blots as log2 fold-change normalized to 45S and the control sample (data average of n = 3 independent experiments; 41S: P = 0.0004, 30S: P = 0.017, 21S: P = 0.027). Hybridization against 45S, and mature 18S and 28S rRNA were used as internal molecular markers. b Immunoprecipitation (IP) with anti-GFP was performed on lysates of cells transfected with constructs expressing either GFP (GFP), FLAG-RRP7A-GFPWT (WT) or FLAG-RRP7A-GFPW155C (MUT), and IPs were analyzed by WB using antibodies against NOL6 and NOL12. The experiments were repeated three times, and results from one representative experiment are shown. c IFM analysis on the localization of NOL6 and NOL12 (green) and RRP7A (red) to nucleoli (open arrows, differential interference contrast microscopy, DIC) in cultures of control and patient HDFs. The circumference of the nucleus is marked with a stippled line. Scale bar, 10 µm. d Quantification of the relative levels of NOL6 (ctr = 70 cells, pt = 66 cells, data average of n = 3 independent experiments; P = 5.749E-06) and NOL12 (ctr = 70 cells, pt = 73 cells, data average of n = 3 independent experiments; P = 0.808) in nucleoli in control and patient HDFs shown in c. Data are represented as mean ± SD and significance was determined using an unpaired, two-tailed Student’s t test. *P < 0.05, **P < 0.01, ****P < 0.0001, n.s.: not significant.