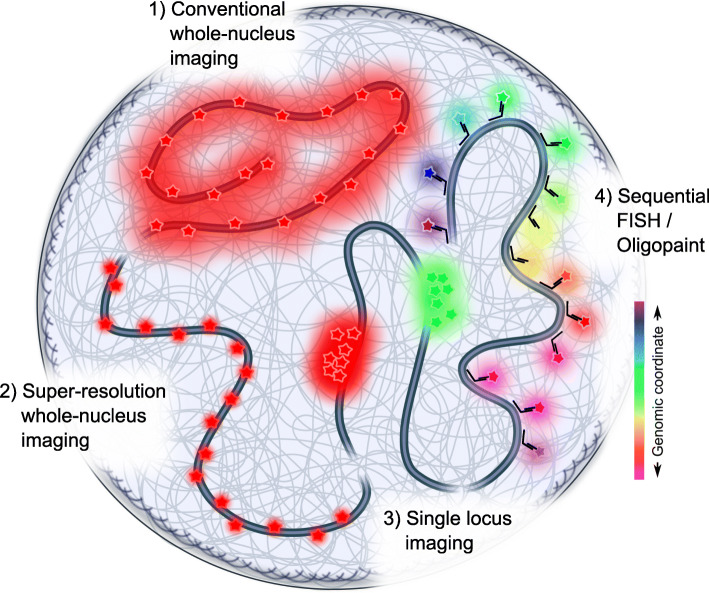
Fig. 2.
Labeling and imaging strategies to image chromatin. Conventional labeling using stably expressed fluorescent proteins or organic dyes usually covers the whole genome unspecifically (modality 1). Due to the high density of chromatin, only a spatial resolution well above the diffraction limit can be achieved, while fast imaging is generally possible. Super-resolution imaging of chromatin overcomes the diffraction limit but is challenging since chromatin in vivo is not well structured (modality 2). Usually, resolution is gained for the expense of acquisition time and thus cells must be fixed. While conventional and super-resolution whole-genome labeling is sequence-unspecific, single loci can be targeted using fluorescence in situ hybridization (FISH; in case of fixed cell imaging), ANCHOR, a CRISPR/Cas9 system, or others (modality 3). Single locus labeling can be done in fixed as well as in living cells. To cover an extended region of chromatin for which sequence information is available, a sequential FISH or oligopaint strategy can be followed in fixed cells (modality 4). Using oligonucleotides which are designed for the specific genomic region, the chromatin configuration is sampled by sequential hybridization rounds and computationally reconstructed