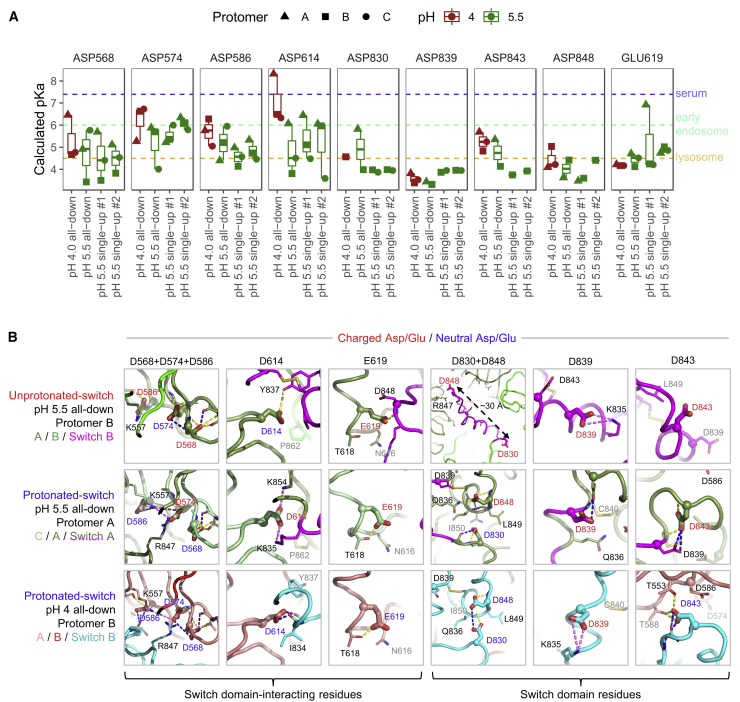
Figure 6.
pKa Calculations for the pH-Switch Domain
(A) PROPKA-calculated pKas for pH-dependent switch domain residues in the pH 4.0 and 5.5 unliganded spike structures. pKas are plotted for titratable residues within and interacting with the 824–858 pH-dependent switch domain for in each structure, disordered regions excluded. Typical pH values for serum (7.4), early endosome (6.0), and late endosome (4.5) are indicated by dashed lines.
(B) Close-up views of Asp/Glu residues in (A) from the pH 4.0 and pH 5.5 structures depict changes in chemical environment for each residue between conformations. View angles with respect to superposed structures are the same within each residue column. Switch domain and surrounding protomers are colored as indicated at left. Highlighted residues are shown as thick sticks with labels colored based on pKa-based dominant protonation state at the structure pH: charged Asp/Glu in red, and neutral (protonated) Asp/Glu in blue. Residues within 4 Å are shown as thin sticks. Dashed lines indicate hydrogen bonds (yellow) or salt bridge interactions (violet), and hydrogen bonds requiring carboxylic acid group protonation are shown in blue. The pKa shifts between unprotonated- and protonated-switch conformations define a pH-dependent stability gradient that favors the protonated-switch form at lower pHs (Yang and Honig, 1993). However, other factors such as global conformational constraints might also play a role in favoring one conformation over another.