Figure 2.
Target Prediction Analysis Identified Shared Pathways among ACE2 Regulators
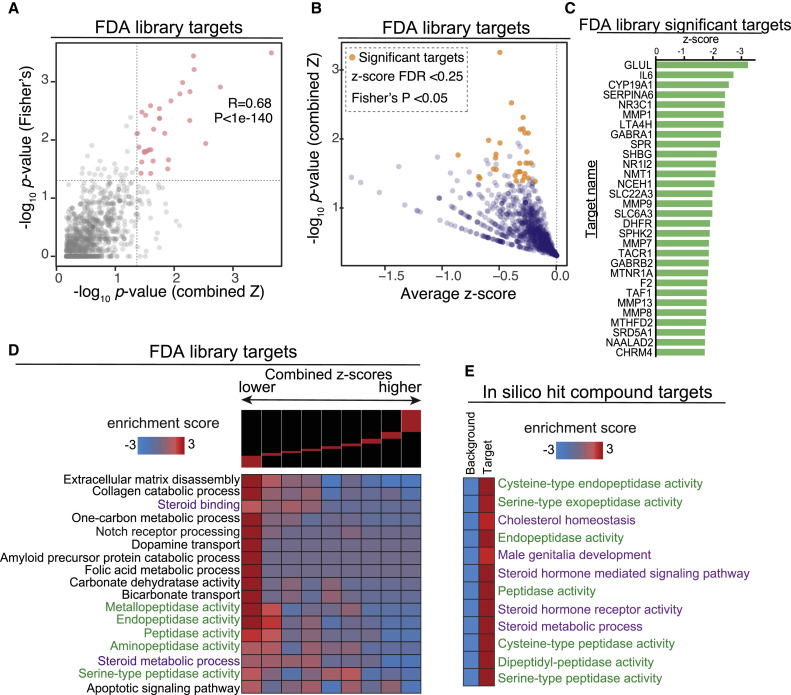
(A) We employed two independent tests for identifying the genes that are most likely targeted by the effective treatments: (1) a combined z-score approach, where normalized z-scores from all the treatments associated with a gene are integrated, and (2) a Fisher’s exact test to assess the enrichment of a gene among those that are targets of treatments with negative z-scores. Here we have shown the correlation between the p values reported by these two independent approaches.
(B) A one-sided volcano plot showing the average z-score versus −log of p value for all genes with negative z-scores. The genes that pass our statistical thresholds are marked in gold (combined z-score FDR < 0.25 and Fisher’s p value < 0.05).
(C) The identified target genes along with their combined z-score, associated p value, and FDRs. Also included are the total number of compounds each gene is likely targeted by, and the number of those that result in lower ACE2 expression (z-score < 0).
(D) Gene-set enrichment analysis using iPAGE for the target genes identified from FDA-approved library with negative z-score. Genes were ordered based on their combined z-score from left to right and divided into nine equally populated bins. The enrichment and depletion pattern of various gene-sets is then assessed across this spectrum using mutual information. Red boxes show enrichment and blue boxes show depletion.
(E) Gene-set enrichment analysis for the in silico hits. Similar to (D), genes were grouped into those that are likely targeted by the identified compounds and those that are not (i.e., background). We then assessed the enrichment of each pre-compiled gene-set among the targets using iPAGE.