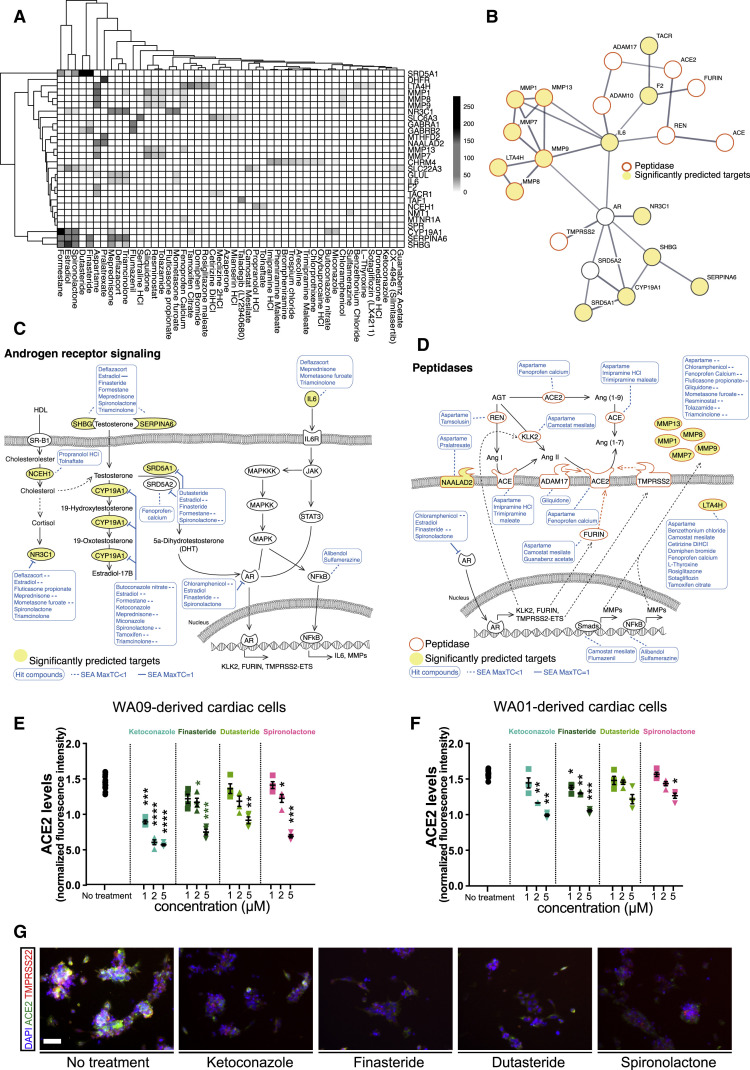
Figure 3.
Androgen Signaling Regulates Peptidase Expression
(A) The drug-gene interaction matrix for the 30 significantly enriched drug target genes from Figure 2C that are deemed functional in their respective analyses. Shading represents the significance of the predicted interaction.
(B) STRING protein-protein interaction network was used to identify interactions between our list of significantly enriched genes from Figure 2C (depicted as significantly predicted targets and yellow circles), androgen signaling pathway components (AR and SRD5A2), and proteins implicated in ACE2 regulation (ACE, ADAM10, ADAM17, FURIN, REN, TMPRSS2). Minimum required interaction score was set to 0.7 corresponding to high confidence and edge thickness indicates the degree of data support.
(C) SEA predicted drug-protein target interactions (blue lines and boxes) in the androgen signaling pathway. Yellow ovals represent significantly enriched genes from Figure 2C. Dashed lines represent MaxTC < 1.
(D) The expression of ACE2-related peptidases is regulated by AR and other transcription factors that are targets of our candidate drugs. MaxTC, maximum tanimoto similarity between compounds from ref_target to compounds from query_target in [0,1] with 1 being identical up to the resolution of the fingerprint.
(E and F) Dose response analysis of the effects of antiandrogenic drug candidates on ACE2 expression in cardiac cells generated from hECS lines WA09 (E) and WA01 (F). Data are represented as mean ± SEM.
(G) Representative images of immunofluorescence staining for ACE2 and TMPRSS2 in hESC-derived cardiomyocytes treated with antiandrogenic drugs. Scale bar = 50 μm.
∗p value < 0.05, ∗∗p value < 0.01, ∗∗∗p value < 0.001. See also Figure S3 and Table S3.