Figure 28.
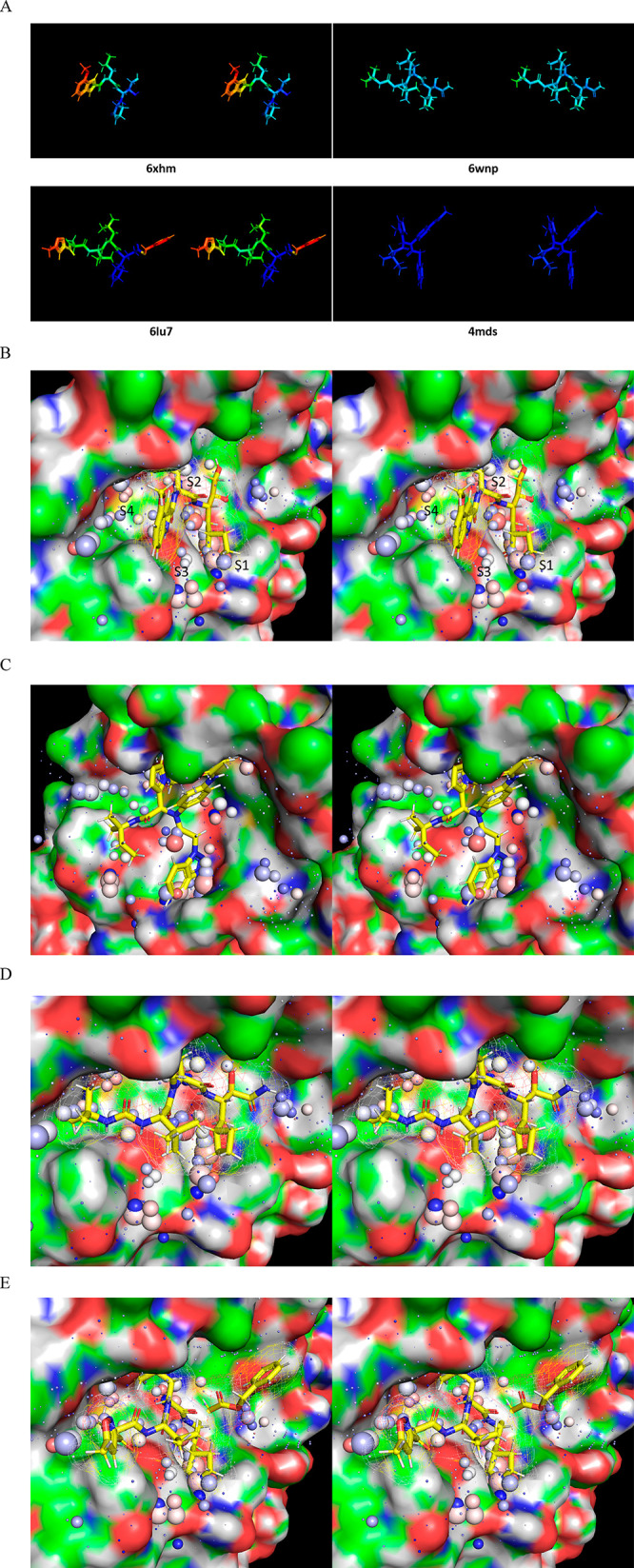
Stereo views of four representative crystallized inhibitors overlaid on the time-averaged M1/downpro structure (2QCY) and the solvation structure thereof calculated using WATMD. ULOVs are distributed diffusely across the S1′ through S4 subpockets, each of which additionally contain clusters of HOVs representing one or two water molecules per cluster (noting that the sphere sizes are proportional to occupancy, rather than the spatial expanse of the voxels). Inhibitor-solvation structure complementarity assessment is based on overlaps between polar/nonpolar inhibitor R-groups and ULOVs, together with overlaps between polar/nonpolar R-groups and HOVs (acceptors with red to pink HOVs; donors with blue to light blue HOVs; and no overlaps between HOVs and nonpolar groups). Complementarity between the inhibitor R-groups and HOVs is outlined in the text and Supporting Information. (A) B-factors of the crystallized inhibitors bound to Mpro. (B) 6XHM/PF00835321 (Ki = 0.27 nM).29 (C) 4MDS/SID 24808289 (IC50 = 6.2 μM, noting the existence of a 51 nM analog 17a).30 (D) 6WNP/boceprevir (IC50 = 8 μM).41 (E) 6LU7/N3 (IC50 = 125 μM).26
