Table 1.
Antifiloviral Activity of 4-(Aminomethyl)benzamides 5-8and 10-27.
| N | Structure | Ebola pseudovirus A549 cells | Marburg pseudovirus A549 cells | |||
|---|---|---|---|---|---|---|
| % inhibition | EC50a (μM) | SIb | % inhibition | EC50a (μM) | ||
 | ||||||
| 5 | X = CH2; para- | 74.0 | 9.86 ± 2.14 | 15 | 90.7 | 0.53 ± 0.09 |
| 6 | X = O; para- | 57.2 | - | 68.8 | - | |
| 7 | X = N-Me; para- | 29.4 | - | 30.3 | - | |
| 8 | X = CH2; meta- | 0 | - | 0 | - | |
| 9 | 13.0 | - | 5.90 | - | ||
| 10 | 12.9 | - | 0 | - | ||
 | ||||||
| 11 | R = 4-CO2H | 3.12 | - | 10.3 | - | |
| 12 | R = 4-SO2Me | 0 | - | 17.4 | - | |
| 13 | R = 4-SO2NHMe | 0 | - | 3.30 | - | |
| 14 | R = 4-NHSO2Me | 3.91 | - | 6.81 | - | |
| 15 | R = 4-CN | 0 | - | 12.4 | - | |
| 16 | R = 4-CF3 | 89.9 | 3.87 ± 0.73 | 9 | 79.3 | 5.70 ± 1.56 |
| 17 | R = 3-CF3 | 82.2 | 2.97 ± 0.20 | 15 | 57.5 | 1.41 ± 0.32 |
| 18 | R = 3-Cl, 4-CF3 | 99.7 | 2.34 ± 0.57 | 9 | 98.5 | 2.05 ± 0.29 |
| 19 | R = 3-CF3, 4-Cl | 99.6 | 1.52 ± 0.28 | 7 | 98.2 | 1.24 ± 0.24 |
| 20 | R = 3-Cl, 4-SCF3 | 99.9 | 1.27 ± 0.38 | 5 | 99.8 | 1.61 ± 0.15 |
| 21 | R = 4-cyclo-Pr | 74.3 | 4.57 ± 1.50 | 22 | 66.2 | 2.65 ± 1.64 |
| 22 | R = 4-i-Pr | 93.1 | 2.87 ± 0.38 | 10 | 67.5 | 2.10 ± 1.24 |
| 23 | R = 2,4-di-t-Bu | 99.5 | 0.48 ± 0.17 | 76 | 85.0 | 2.34 ± 0.64 |
| 24 | 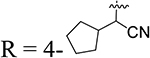 |
96.8 | 0.73 ± 0.29 | 19 | 94.0 | 1.63 ± 0.42 |
| 25 | R1 = R2 = H | 98.1 | 0.99 ± 0.18 | 101 | 73.4 | 0.96 ± 0.32 |
| 26 | R1 = F, R2 = H | 96.1 | 0.38 ± 0.12 | 382 | 71.0 | 1.33 ± 1.23 |
| 27 | R1 = H, R2 = Me | 99.3 | 0.33 ± 0.08 | 390 | 88.9 | 0.85 ± 0.10 |
| toremifene | 0.09 ± 0.04 | 299 | ||||
Dose-response studies were conducted to determine EC50 values for those compounds that showed more than 70% inhibition at 12.5 uM concentration; EC50 values were calculated by four-parameter dose-response curve-fitting in GraphPad. Results are from three replicates. Percent inhibition errors are estimated to be <10%; EC50 data are presented as mean ± standard deviation (SD).
Selectivity index is the ratio of CC50/EC50, where CC50 is cytotoxicity assessed by utilizing the “CellTiter 96 aqueous nonradioactive cell proliferation assay” (Promega, Madison, WI).
