Figure 1.
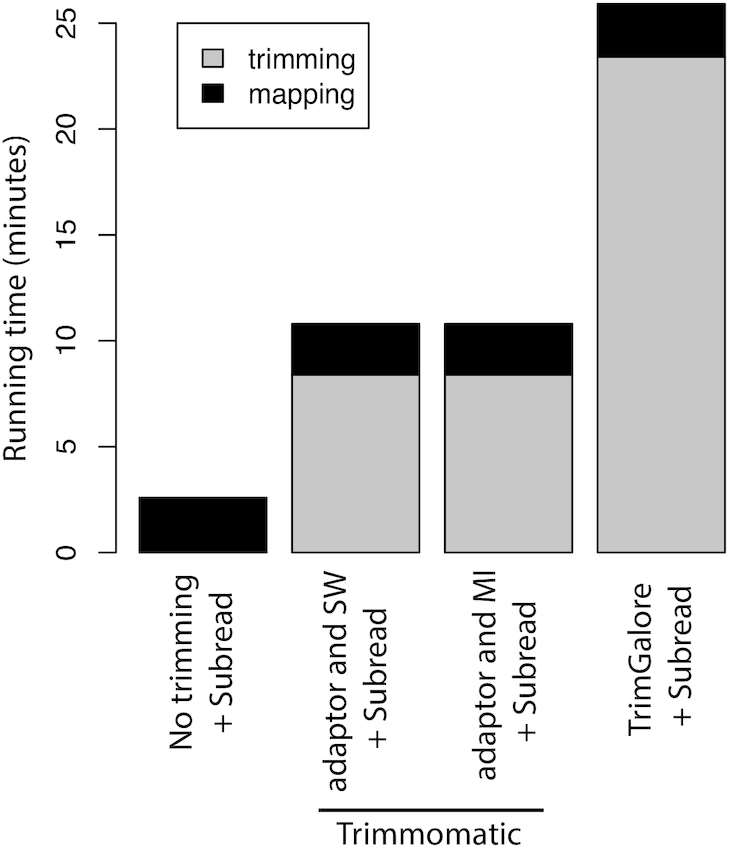
Time cost of different methods running on a UHRR RNA-seq dataset that includes 15 million 100 bp read pairs. All software tools were run with eight CPU threads. Input data to trimming and mapping tools are in gzipped FASTQ format which is the standard format of data generated by Illumina sequencers.
