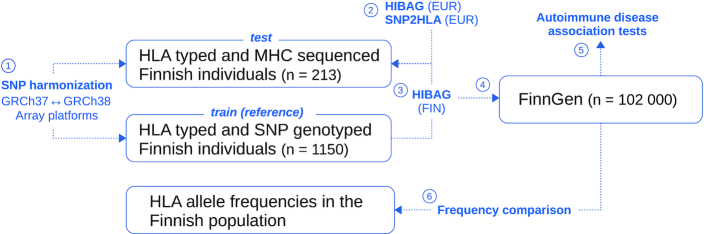
Figure 1.
Schematic diagram of the analysis steps. (1) MHC SNPs were selected for consistency between array platforms and genome builds. The SNP genotype data with clinical-grade HLA types were divided into training and test sets. (2) HIBAG and SNP2HLA programs with their default references were applied to the test set to compare their accuracies. (3) HIBAG models were built on the training set and applied to the test set to analyze the impact of population-specific reference data on imputation accuracy. (4) HIBAG models trained on the Finnish reference data were applied to FinnGen R2 cohort. (5) The imputed HLA were tested for possible associations with six common autoimmune disorders to replicate known risk alleles. (6) Frequencies of the imputed HLAs were compared with frequencies from an independent clinical-grade HLA dataset.