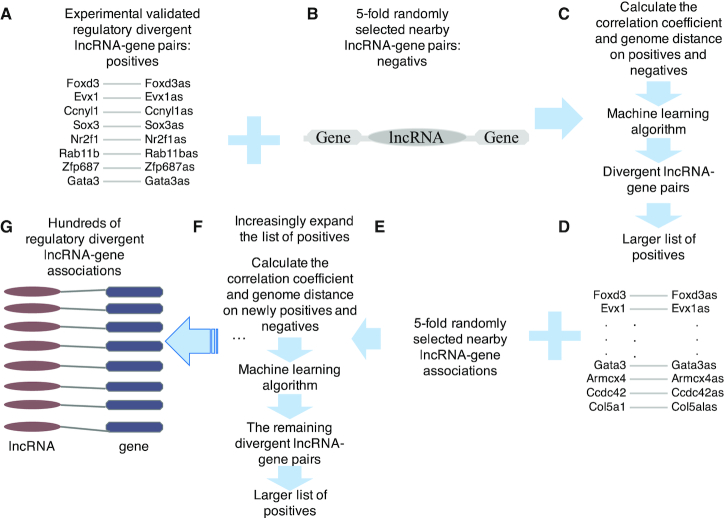
Figure 2.
The increasingly learning framework. (A) The training positives: experimentally validated divergent lncRNA and gene regulations. (B) The training negatives: 5-fold randomly selected nearby lncRNA-gene pairs. (C) The machine learning algorithm that is taken validated regulatory lncRNA-gene pairs as training positives, 5-fold randomly selected nearby lncRNA-gene pairs as the training negatives and divergent lncRNA-gene pairs as the testing set, was introduced to generate the larger list of positives. The genome sequence, expression and chromatin accessibility correlation coefficients and genome distance were used as input features for that machine learning algorithm. (D) The increased regulatory divergent lncRNA-gene pairs generated from the machine learning algorithm. (E) Another 5-fold randomly selected nearby lncRNA-gene pairs. (F) The iterative process to generate the larger list of positives with genome sequence, expression and chromatin accessibility correlation coefficients and genome distance as the input feature for newly positives and negatives. (G) Hundreds of regulatory divergent lncRNA-gene associations generated via increasingly expanding the list of positives.