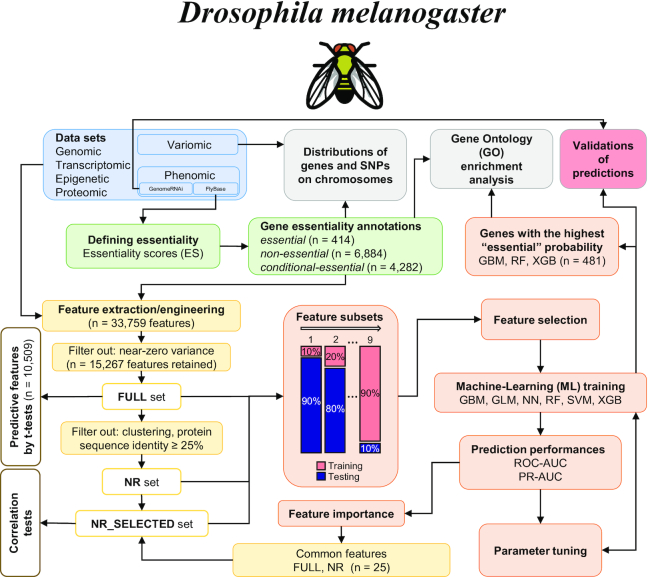
Figure 1.
Workflow employed in the present study. First, a wealth of publicly available ‘omics datasets for Drosophila melanogaster were obtained (blue). Then, we employed a ‘scoring system’ to annotate D. melanogaster genes for essentiality (green) using phenomic data. Next, we extracted or engineered features (yellow) from the datasets to establish feature sets (FULL—all features; NR—all features from sequences containing <25% amino acid identity; NR_SELECTED—25 highly predictive features of essentiality, selected from the NR dataset). These feature sets were used for a systematic evaluation of ML approaches for essential gene predictions (orange). Statistical significance (t-tests) and correlation tests were performed on the FULL and NR_SELECTED sets, respectively. The performances of the individual ML models, and the importance of the selected features for essentiality predictions were calculated and evaluated (orange). Independent validations of the ML predictions using knockdown (RNAi) data was also performed (red). Finally, GO enrichment and preferential genomic locations of SNPs and genes by essentiality annotations were evaluated (gray).