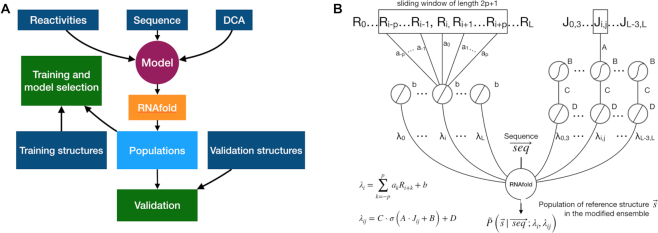
Figure 1.
Graphical scheme of the machine learning procedure. (A) Models that integrate RNAfold, chemical probing experiments and DCA scores into prediction of structure populations are trained. One among all the proposed models is selected based on a transferability criterion and validated against data that is not seen during training. Available reference structures are used as target for training and validation. (B) Sequence, reactivity profile and DCA data are included through additional terms in the RNAfold model free energy. The network is split into two channels: a single-layered channel for reactivity input (left side) and a double-layered channel for DCA couplings (right side). Along the reactivity channel, a convolutional layer operates a linear combination on the sliding window including the reactivity Ri of a nucleotide and the reactivities {Ri + k} of its neighbors, with weights {ak} and bias b. The output consists in a pairing penalty λi for the i-th nucleotide. In the DCA channel, the first layer transforms the input DCA coupling Jij via a non-linear (sigmoid) activation function, with weight A and bias B. The transformed DCA input is then mapped to a pairing penalty λij for the specific ij pair via a second layer, implementing a linear activation function with weight C and bias D. Penalties for both individual nucleotides and for specific pairs are applied as perturbations to the RNAfold free-energy model.