Table 1.
Run time in seconds for different alignment-free approaches on six sets of real-world genomes. On the largest data set, the 14 plant genomes, ‘kmacs’ and ‘co-phylog’ did not terminate the program run. On this data set, we increased the pattern weight for ‘Multi-SpaM’ from the default value of  = 10 to
= 10 to 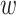 = 12, in order to reduce the run time. Note that ‘Multi-SpaM’, ‘FSWM’ and ‘andi’ are parallelized, so we could run them on multiple processors, while ‘kmacs’ and ‘co-phylog’ had to be run on single processors. The reported run times are ‘wall clock times’.
= 12, in order to reduce the run time. Note that ‘Multi-SpaM’, ‘FSWM’ and ‘andi’ are parallelized, so we could run them on multiple processors, while ‘kmacs’ and ‘co-phylog’ had to be run on single processors. The reported run times are ‘wall clock times’.
| FSWM | andi | co-phylog | kmacs | Multi-SpaM | ||||
|---|---|---|---|---|---|---|---|---|
| wall clock | CPU | wall clock | CPU | wall clock | CPU | |||
| 27 E.coli/Shigella | 710 | 27,075 | 15 | 291 | 704 | 5,247 | 603 | 22,185 |
| 29 E.coli/Shigella | 860 | 32,798 | 16 | 325 | 533 | 55,736 | 611 | 21,973 |
| 25 fish mitochondria | 2 | 8 | <1 | 3 | 9 | 5 | 27 | 1,054 |
| 14 plants | 1,107,720 | 28,690,489 | 1,808 | 13813 | - | - | 12,516 | 389,770 |
| 19 Wolbachia | 65 | 2,185 | 3 | 42 | 113 | 24,961 | 484 | 15,804 |
| 8 Yersinia | 91 | 3,333 | 5 | 34 | 50 | 1,083 | 183 | 6,182 |
