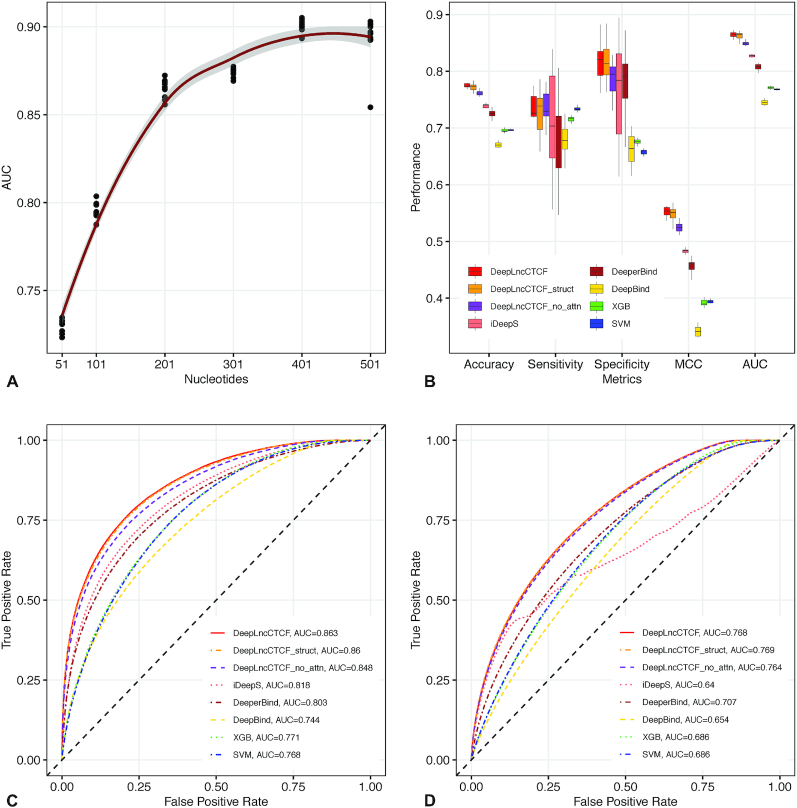
Figure 2.
Accurate prediction of CTCF-binding RNA sites by DeepLncCTCF. (A) The AUC of DeepLncCTCF using six different input sequence lengths in nucleotides. The trend line was fitted using polynomial regression. The shaded area indicates the 95% confidence intervals. (B) Comparison of model performance based on accuracy, sensitivity, specificity, MCC and AUC using the high-confidence test dataset of human U2OS cells. The models are DeepLncCTCF, DeepLncCTCF_struct (using both RNA sequences and secondary structure information as input), DeepLncCTCF_no_attn (without the attention layer), iDeepS (41), DeeperBind (40), DeepBind (37), XGB and SVM. (C) ROC curves of the models on the high-confidence test dataset of human U2OS cells. (D) ROC curves of the models on the separate, low-confidence test dataset of human U2OS cells (see the ‘Materials and Methods’ section).