Figure 4.
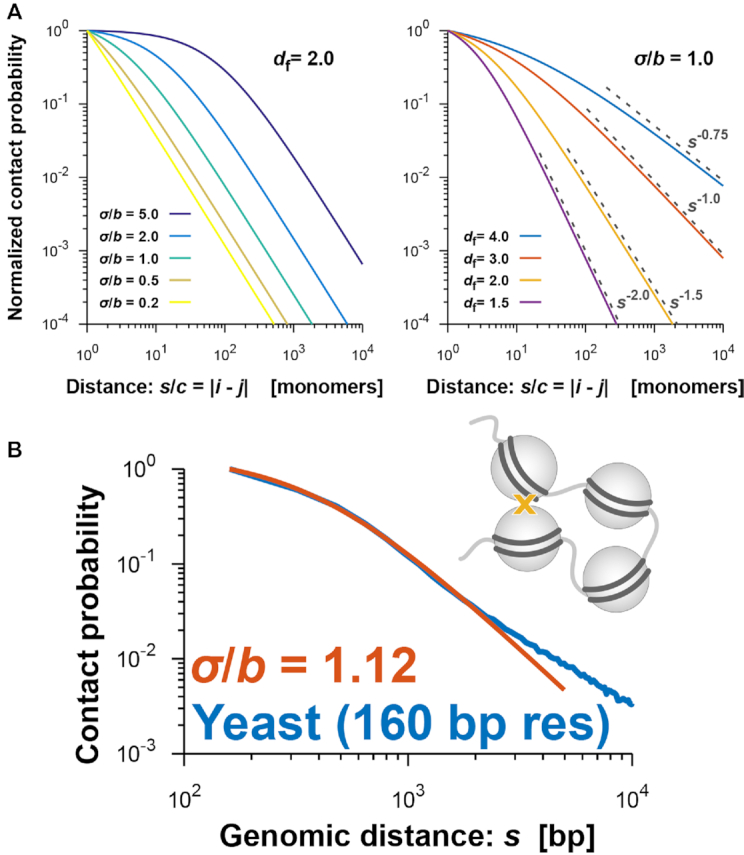
(A) Theoretical curves of the contact probability P(s) in Equation (4). P(s) is normalized by the value at s/c = |i − j| = 1, where i and j are monomer indices of the fractal polymer and c represents the genomic size corresponding to every monomer. (Left) P(s) for fixed df = 2.0 and σ/b = 0.2, 0.5, 1.0, 2.0 and 5.0. (Right) P(s) for fixed σ/b = 1.0 and df = 1.5, 2.0, 3.0 and 4.0. Theoretical scaling relations, as in Equation (5), are shown. (B) Contact probability (blue) for yeast cells with 160-bp nucleosome resolution (31) as a function of genomic distance averaged across the genome, and the theoretically fitted curve (orange) at a small genomic distance.
