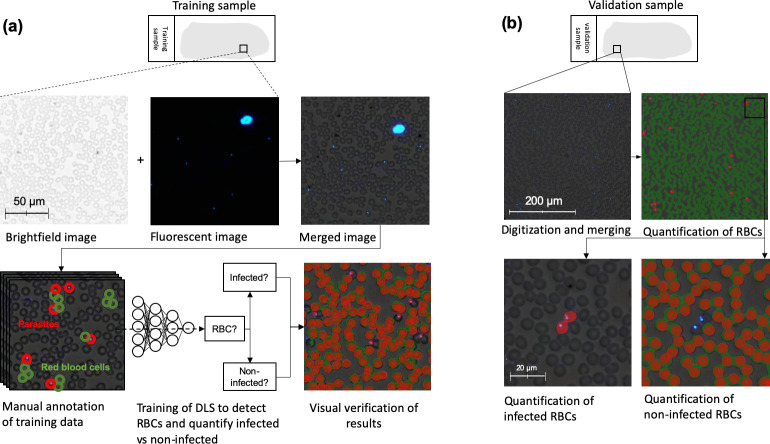
Fig 3. Training of deep-learning system 1 and analysis of blood smear.
Workflow for training of the first deep-learning system (DLS 1) and subsequent analysis of samples using the trained model. (a): Training of the DLS was performed on merged images (brightfield and fluorescent images combined) where regions with visible red blood cells (RBCs) and trophozoites (parasites) were manually annotated and used to train the DLS to detect RBCs and classify them as infected vs. non-infected. (b): Analysis of samples in the validation series was performed on merged images in two steps: 1) Segmentation of all visible RBCs and 2) quantification of infected and non-infected RBCs to determine overall level of parasitaemia.