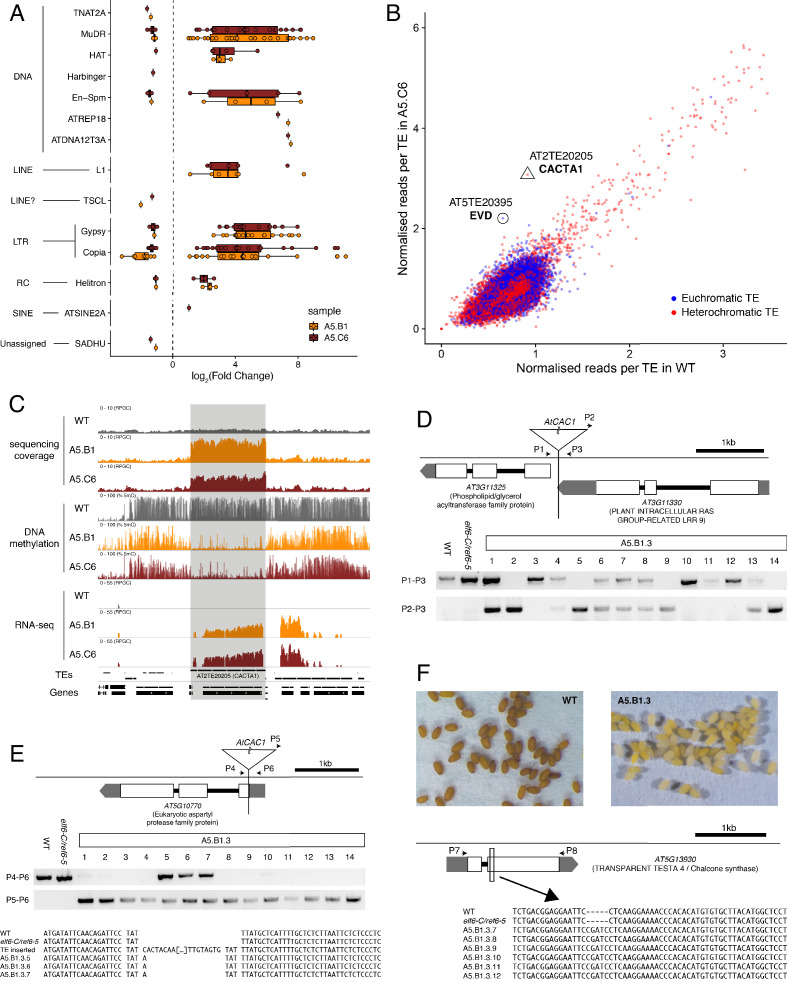
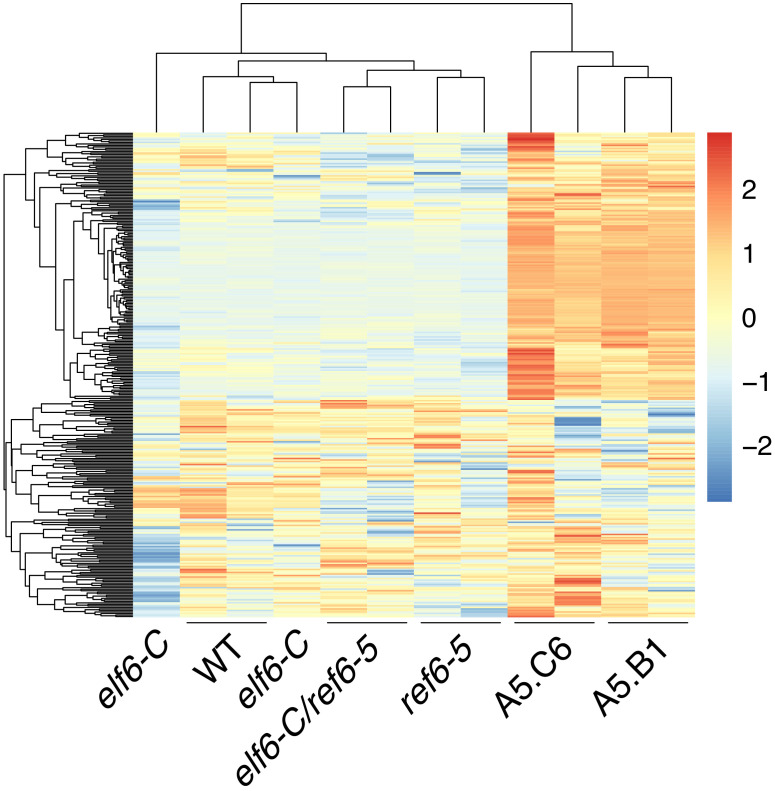
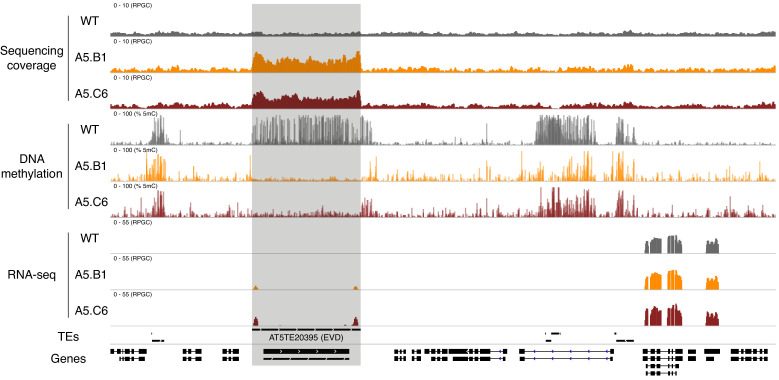
Figure 6. Transposon mobilization in epiERs results in heritable genetic lesions.
(A) Differential expression of DNA and RNA transposon families grouped by superfamily in progenies of epiERs A5.B1 and A5.C6. (B) Copy number variation of transposons in progenies of epiER A5.C6. Blue dots, euchromatic TEs; Red dots, heterochromatic TEs. (C) Genome browser views of normalized sequencing coverage (RPGC), DNA methylation frequency (%) and RNAseq coverage (RPGC) in wild-type (WT) and progenies of epiERs A5.B1 and A5.C6. Grey box, AT2TE20205 (CACTA1). (D) Map of transposon insertion in AT3G11330 and its segregation in epiER A5.B1.3 progenies. P1-3, primers used for PCR amplification and sequencing. (E) Map of transposon insertion in AT5G10770 and sequence footprint resulting from re-mobilization in epiER A5.B1.3 progenies. P4-6, primers used for PCR amplification and sequencing. (F) Seed pigmentation defects caused by a sequence insertion in AT5G13930 (TRANSPARENT TESTA4/CHALCONE SYNTHASE) resulting from transposon re-mobilization in in epiER A5.B1.3 progenies. P7-8, primers used for PCR amplification and sequencing.