Figure 2. Evolution of KAI2 in Parasitic Plants.
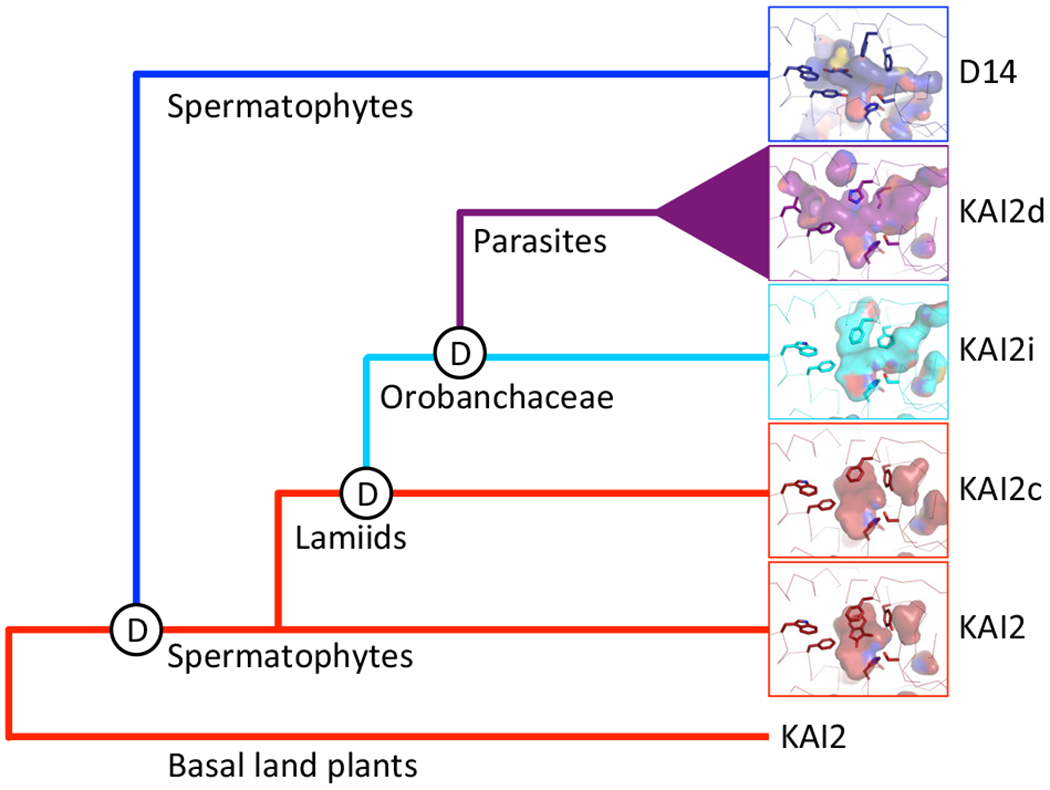
D14 is an ancient paralog of KAI2 that has SL specificity. Duplication of KAI2 in the Lamiid lineage likely gave rise to KAI2c and KAI2i paralogs. Further, extensive duplication of KAI2 in the Orobanchaceae family gave rise to the KAI2d clade found in parasitic plants. The ligand-binding pockets of rice D14 (top, blue) and Arabidopsis KAI2 (bottom, red) are shown with key pocket residues and shading of the hydrophobic cavities. Homology modeling of KAI2 paralogs in parasitic plants indicates merging of two cavities in KAI2i and an increase in ligand-binding pocket size in the KAI2d clade. Similar to D14, KAI2d proteins recognize SLs, suggesting convergent evolution. KAI2i respond to KARs and KAI2c may recognize KL; Arabidopsis KAI2 has both properties, suggesting subfunctionalization may have occurred in the Lamiids. Abbreviations: KAI2, KARRIKIN-INSENSITIVE2; D14, DWARF14; KAR, karrikin; SL, strigolactone; KL, KAI2 ligand.
