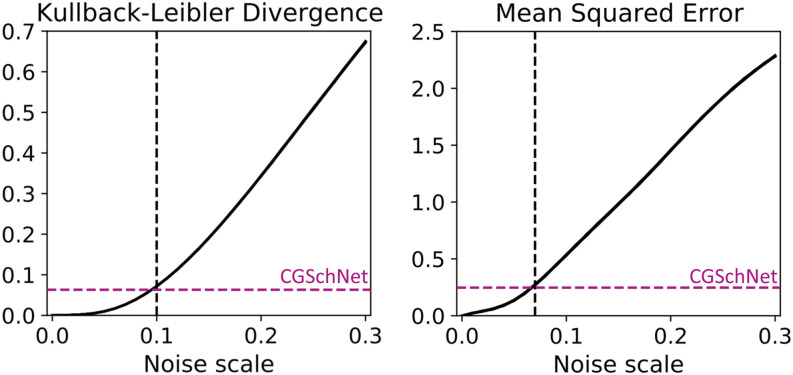
FIG. 5.
The Kullback–Leibler divergence (left) and mean squared error (right) are calculated between the baseline capped alanine dataset and the datasets obtained from perturbations to the baseline simulation at noise scale values of σ ∈ {0, 0.01, 0.02, …, 0.30} where the former is the reference distribution and the latter is the trial distribution. This procedure is performed 50 times with different random seeds; both plots show the superposition of those 50 lines. The colored horizontal dashed line shows the value of the metric when comparing the CGSchNet simulation to the baseline, and the black vertical dashed line indicates the noise scale σ that return the closest value for that metric. The Kullback–Leibler (KL) divergence is computed for normalized bin counts in ϕ × ψ space, and the MSE is computed for the energies of those bins as described in the main text.