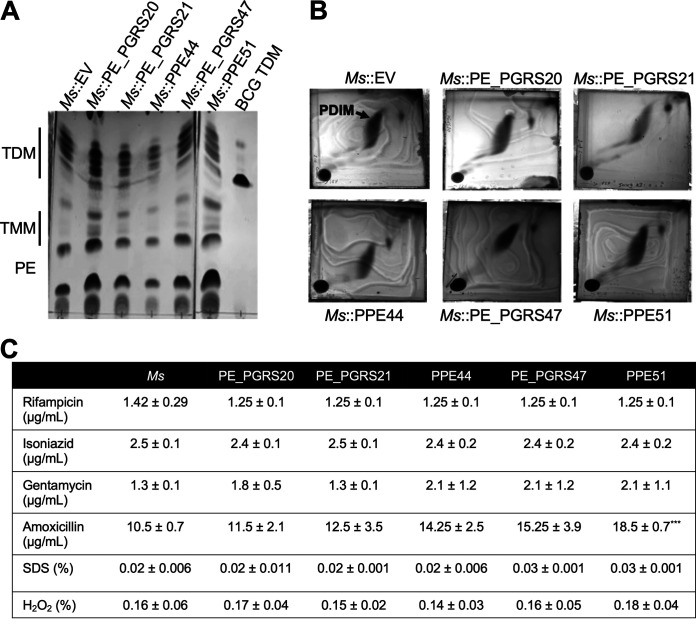
FIG 7.
PE/PPE proteins do not affect M. smegmatis (Ms) lipid composition or antibiotic sensitivity. (A) Thin-layer chromatography of polar lipid extraction from M. smegmatis::empty vector or M. smegmatis::PE/PPE overexpressing strains. TDM, trehalose dimycolate; TMM, trehalose monomycolate; PE, phosphatidylethanolamine; PDIM, phthiocerol dimycocerosates. Polar lipids were analyzed using the solvent system chloroform/methanol/water (20:4:0.5, vol/vol/vol) and visualized by charring with 10% copper sulfate in 8% phosphoric acid. (B) Two-dimensional thin-layer chromatography of apolar lipid extraction from M. smegmatis::empty vector or M. smegmatis::PE/PPE. Apolar lipids were analyzed using the solvent system petroleum ether/ethyl acetate (98:2, vol/vol, 3×) in the first direction and petroleum ether/acetone (98:2, vol/vol) in the second direction. Lipids were visualized by charring with ceric ammonium molybdate in 6% acid. (C) MIC90 of M. smegmatis::empty vector and M. smegmatis::PE/PPE overexpressing strains to various antibiotics as was determined by resazurin reduction assay. The mean ± SD of three independent experiments is shown. Statistical significance was calculated by two-way ANOVA corrected for multiple comparisons by Dunnett’s test. ***, P ≤ 0.001.