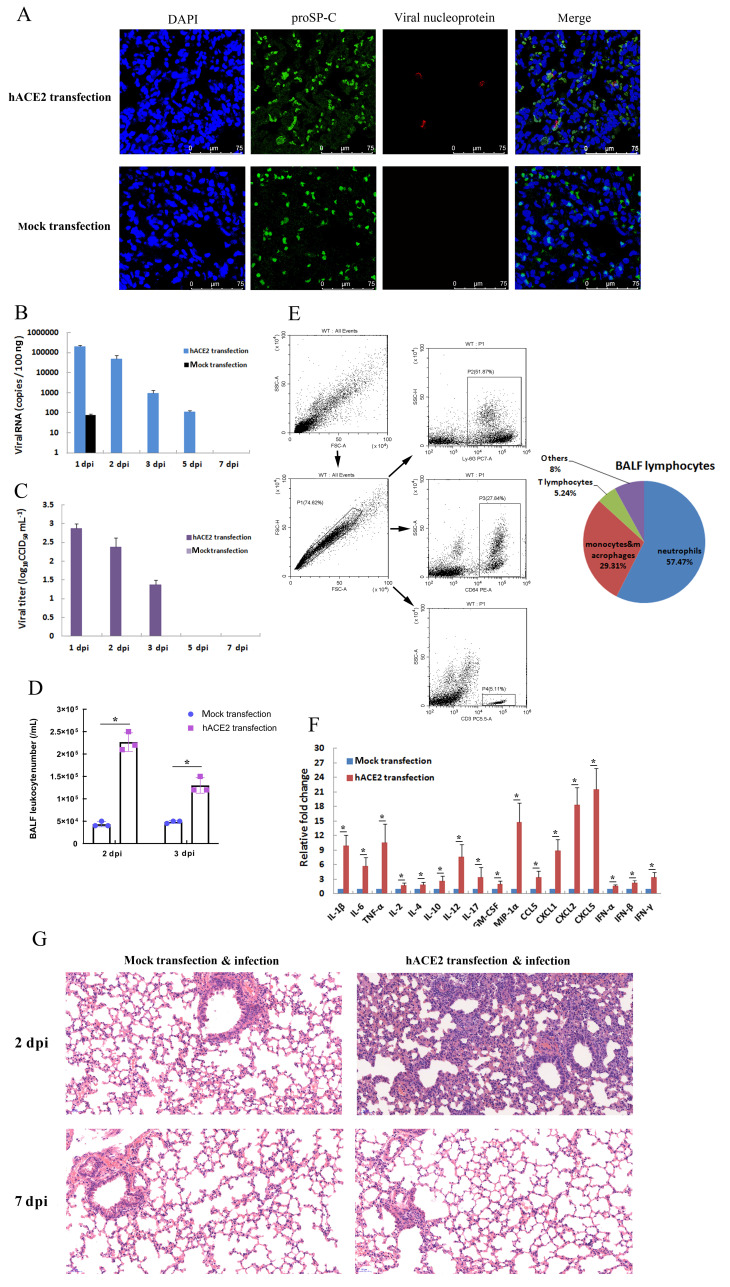
Figure 3.
Pulmonary SARS-CoV-2 infection in hACE2-transfected mice
Two days (48 h) after in vivo transfection with hACE2 plasmid or control plasmid (mock transfection), mice were intranasally infected with SARS-CoV-2 (1×105 CCID50). A: Lungs were harvested from infected mice at 2 days post-infection (dpi), fixed in formalin, and embedded in paraffin. Lung sections were stained with anti-proSP-C and viral nucleoprotein for immunofluorescence detection. B, C: Lungs were harvested at different dpi, viral RNA was determined based on number of nucleocapsid (N) gene RNA copies detected by qRT-PCR (B), and viral titers were determined using a CCID50 assay (C). Error bars indicate standard deviation of triplicate biological samples. D: Total numbers of leukocytes in BALF at different dpi were determined. E: Percentages of neutrophils, monocytes and macrophages, and lymphocytes in BALF of infected hACE2 mice at 2 dpi were analyzed by flow cytometry using anti-Ly6G, anti-CD64, and anti-CD3 antibodies, respectively (left). Statistical analysis of BALF lymphocyte composition was carried out with data from triplicate biological samples (right). F: mRNA levels of cytokines from lung tissue homogenates of infected mice at 2 dpi were examined by qRT-PCR (normalized to β-actin). Error bars indicate standard deviation of triplicate biological samples. *: P<0.05 based on Student’st-test. G: Lungs were harvested from infected mice at 2 dpi, fixed in formalin, and embedded in paraffin. Lung sections were stained with hematoxylin and eosin for histopathological analysis (original magnification, 20×).