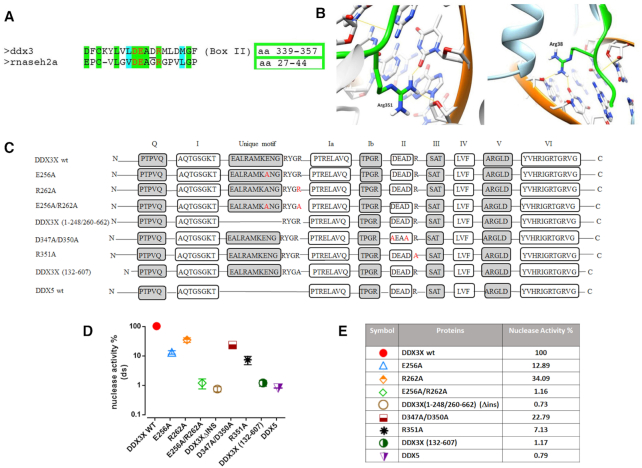
Figure 3.
Identification of DDX3X residues important for its RNaseH2-like activity. (A) Sequence alignment showing amino acid (aa) similarities between the conserved -DEAD- (box II) helicase motif of DDX3X and the catalytic site of human RNaseH2. Red letters indicate catalytically important residues. Green shading indicated identical and cyan shading similar aa. The numbers of the aa residues corresponding to each selected region are boxed. (B) Structure comparison between human DDX3X in open conformation (PDB ID: 5e7I) on the left and mouse RNaseH2 (PDB ID: 3KIO) of the region surrounding Arg351 of DDX3X (left) and Arg38 of RNaseH2 (right), showing the hydrogen bond formed by these residues with the RNA substrate. Arg351 in DDX3X and Arg38 in RNase H2 were similarly positioned within these regions. (C) Graphical representation of all the DDX3X mutants and DDX5 protein. N and C indicate the N-terminal and C-terminal of the proteins respectively. Boxes represent all the conserved motifs of DEAD-box proteins (with the addition of DDX3 unique motif). Red letters indicate single aa mutations. (D) Nuclease activities of the DDX3X mutants and DDX5 protein relative to DDX3X wild type (taken as 100%). All catalytic activities were assessed on Substrate *D39R1D15: D55. Values are the mean of four independent experiments ± S.D. All proteins were tested at 2 μM. (E) Summary table of the catalytic activities shown in panel D.