Figure 2.
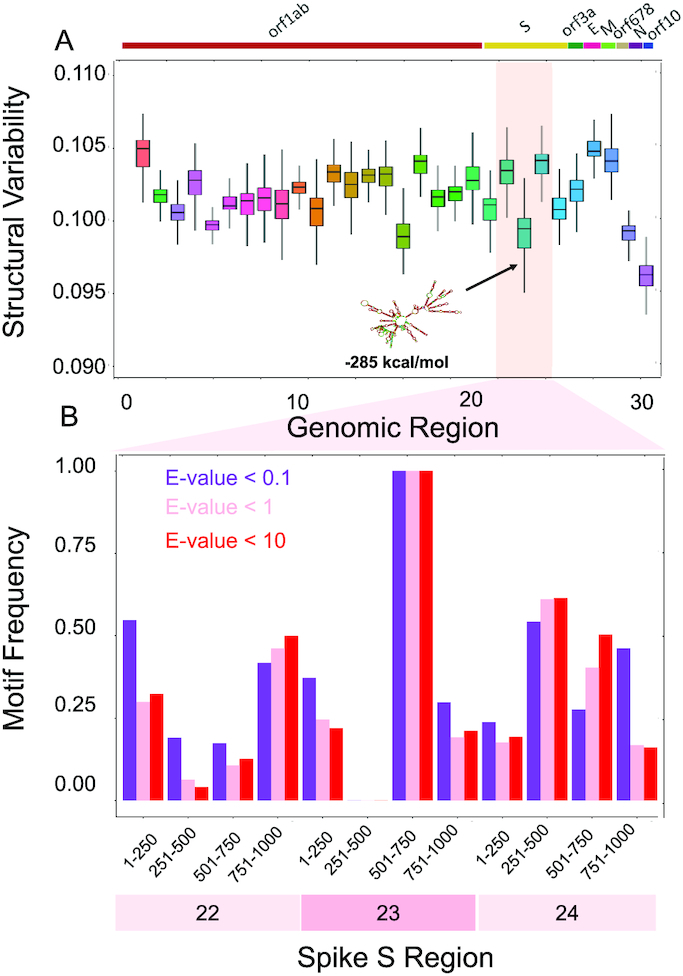
Structural comparisons of coronaviruses. (A) We employed the CROSSalign approach (12,13) to compare Wuhan strain MN908947.3 with other coronaviruses. One of the regions with the lowest structural variability encompasses nucleotides 22 000–23 000. The centroid structure and free energy computed with the Vienna method (25) are displayed. (B) We studied the conservation of nucleotides 22 000–23 000 (fragment 23) and the adjacent regions using structural motives identified with RF-Fold algorithm of the RNAFramework suite (36) with CROSS as soft-constraint. We found that nucleotides 501–750 within fragment 23 are the ones with the highest number of matches at confidence thresholds (E-values).
