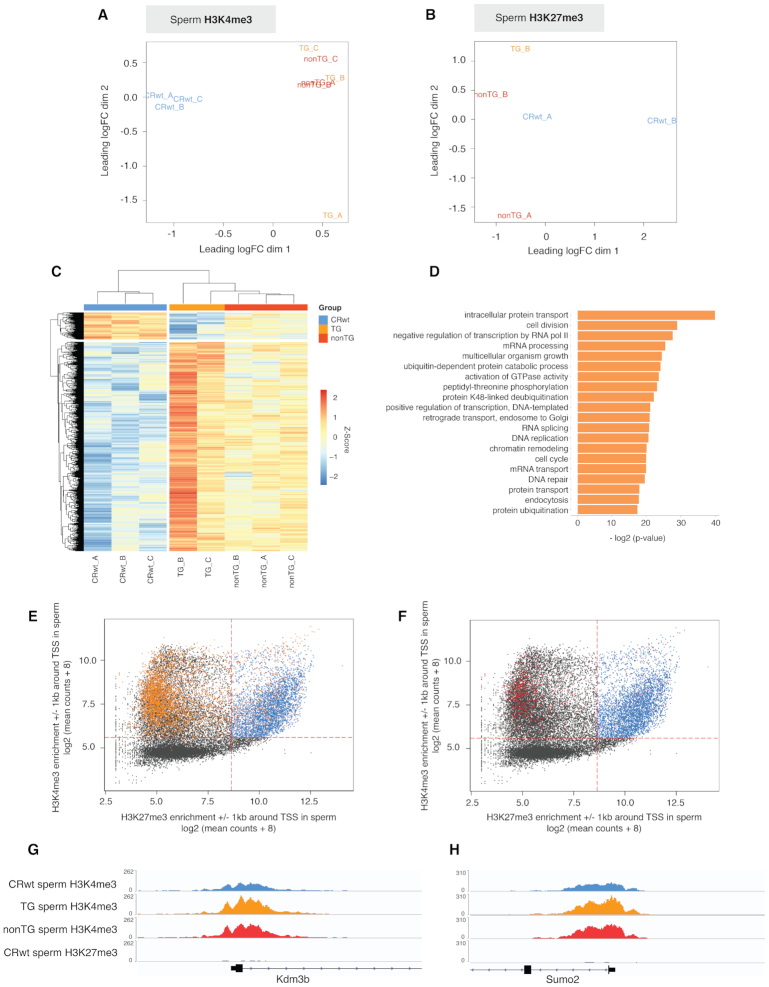
Figure 3.
H3K4me3 is differentially enriched in TG and nonTG sperm at non-bivalent promoters. (A) Multidimensional scaling (MDS) plot of biological replicates for CRwt, TG, and nonTG sperm H3K4me3 normalized windows. TG_A was identified as an outlier and excluded from downstream analysis. (B) MDS plot of biological replicates for CRwt, TG, and nonTG sperm H3K27me3 normalized windows. TG_A failed sequencing (Supplementary Figure S1B) and could not be used for this study. (C) Heatmap of differentially enriched H3K4me3 (deH3K4me3) promoters in TG sperm ( = 3894 deH3K4me3 promoters, FDR < 0.2). (D) Top significant pathways from gene ontology analysis on promoters with deH3K4me3 in TG sperm. (E–F) Scatterplots corresponding to the log2 (mean of CRwt sperm H3K4me3 read counts + 8) ± 1 kb around the TSS of all known genes in the mouse genome against the log2 (mean of CRwt sperm H3K27me3 read counts + 8) ± 1 kb around the TSS, with overlay of regions with deH3K4me3 in TG sperm (orange dots, E) and regions with deH3K4me3 in nonTG sperm (red dots, F). Dashed red lines indicate the count thresholds used to separate bivalent (blue) and non-bivalent (black) chromatin in sperm (see Supplementary Figure S3A and B). Integrative Genome Viewer tracks of (G) the Kdm3b and (H) the Sumo2 promoters, that have deH3K4me3 in TG and nonTG sperm, as well as an absence of H3K27me3 in CRwt sperm.