Figure 6.
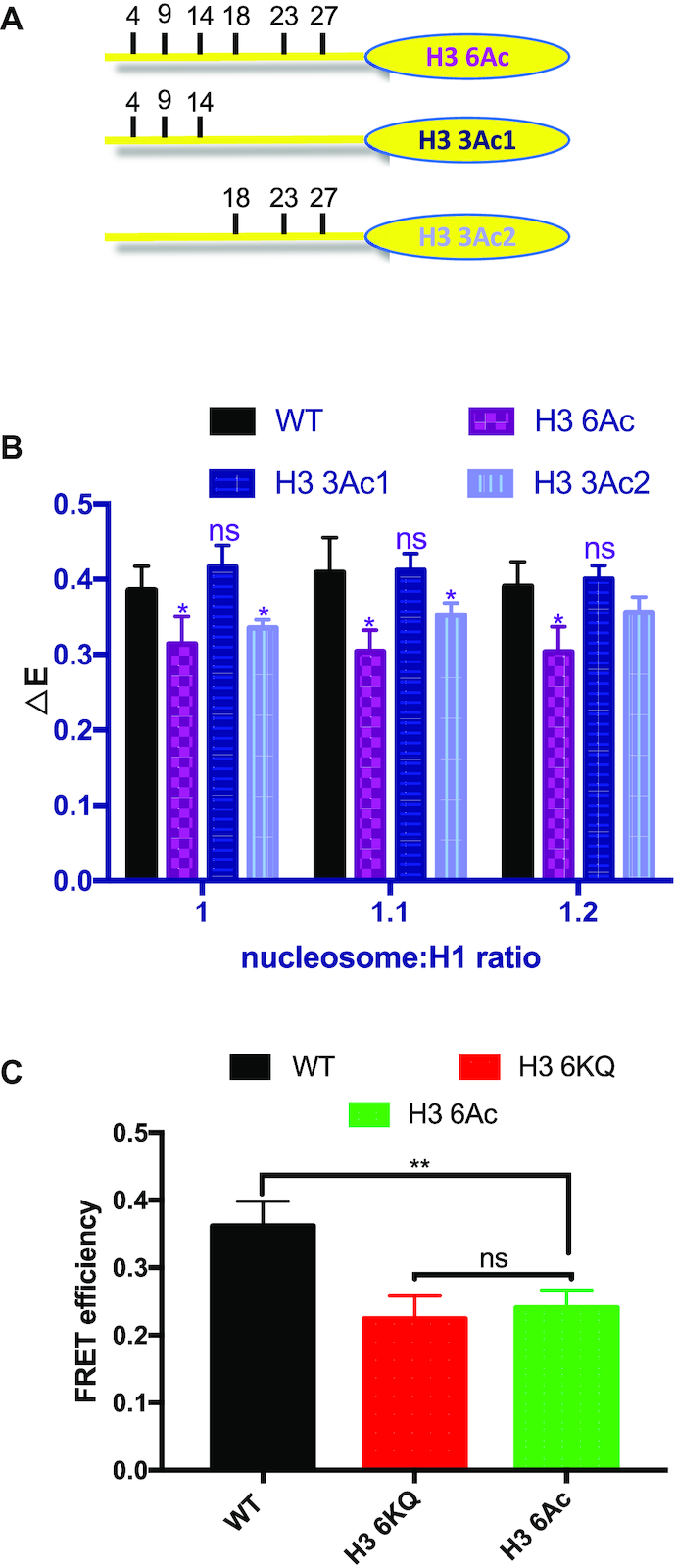
Lysine acetylation in the H3 tail domain affects H1 CTD condensation. H3 proteins containing acetylated lysines at known sites of lysine acetylation were incorporated into nucleosomes and the extent of H1 CTD condensation determined by FRET analysis. (A) Schematic showing sites of lysine acetylation in the H3 tail domain. (B) FRET analysis of labeled H1 incubated with WT and H3 6Ac, H3 3Ac1 and H3 3Ac2 nucleosomes at the indicated nucleosome: H1 ratios. NS, not significantly different from WT; (*) P < 0.05. Note calculated P-value for H3 3Ac2 at ratio 1.2 is 0.07 (not indicated). Analysis of aggregate data for samples in which H1-nuclesome binding is saturated (1.0, 1.1 and 1.2 ratios) indicates P-values of ≤ 3.0E–07, 0.30 and 6.7E-05 for H3 6Ac, H3 3Ac1 and H3 3Ac2, respectively, compared to WT (Supplementary Figure S6). (C) Bona fide lysine acetylation (6Ac) within H3 tail has identical effects on linker DNA end-to-end distance compared to K → Q acetylation mimic (6KQ). NS, not significantly different from WT; (**) P < 0.01.
