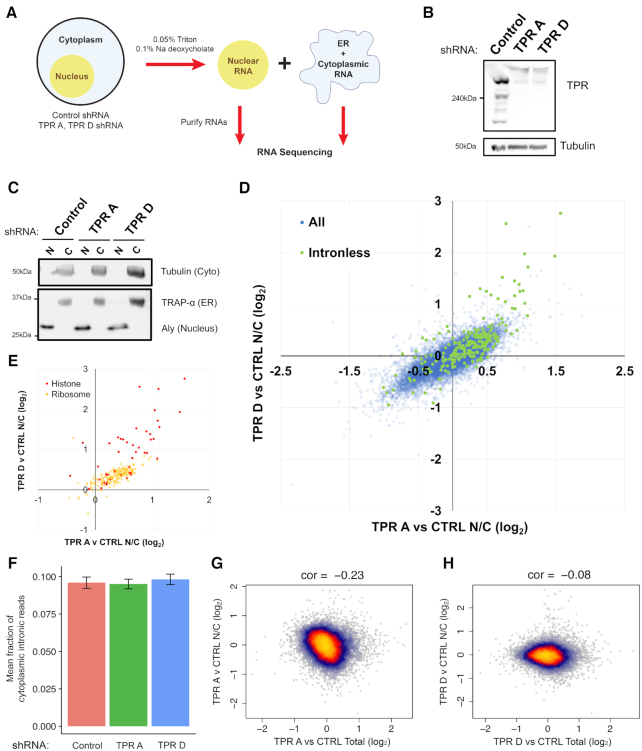
Figure 3.
TPR-depletion inhibits the nuclear export of a subset of endogenous mRNAs. (A) Workflow for RNA Frac-Seq. Control- or TPR-depleted U2OS cells were fractionated into ‘nuclear’ and ‘ER/cytoplasmic’ fractions (see Material and Methods for more details). RNA from each fraction was purified and sent for RNA sequencing. (B) Immunoblot showing TPR-depletion for at least 96 h post-infection using two different lentiviral delivered shRNAs, ‘TPR A’ or ‘TPR D’. Tubulin is used as a loading control. (C) Immunoblot of nuclear ‘N’ or cytoplasmic ‘C’ fractions, probed with specific nuclear (Aly), ER (Trap-α) and cytoplasmic (Tubulin) protein markers. (D, E) Plot of the Log2 fold change of the nuclear/cytoplasmic ratio (TPR v CTRL N/C fold change) between TPR- and control-depleted cells for shRNA ‘TPR A’ (x-axis) or shRNA ‘TPR D’ (y-axis) for each mRNA. Positive values indicate an increase in the nuclear/cytoplasmic ratio in the TPR-depleted cells. All mRNA transcripts are shown in (D) with those transcribed from intronless genes (in green) being more nuclear enriched following TPR-depletion compared to those transcribed from other genes (in blue). Particular classes of mRNAs that have high nuclear enrichment upon TPR-depletion are shown in (E), including histone (in red) and ribosomal (in yellow) mRNAs. (F) Fraction of reads that map back to intronic regions of the genome are shown for cytoplasmic fractions from control and TPR-depleted cells. (G, H) Plot of the TPR-dependent changes in mRNA level (x-axis) versus TPR-dependent changes in nuclear export (TPR v CTRL N/C fold change) for shRNA ‘TPR A’ (G) or shRNA ‘TPR D’ (H) for each mRNA.