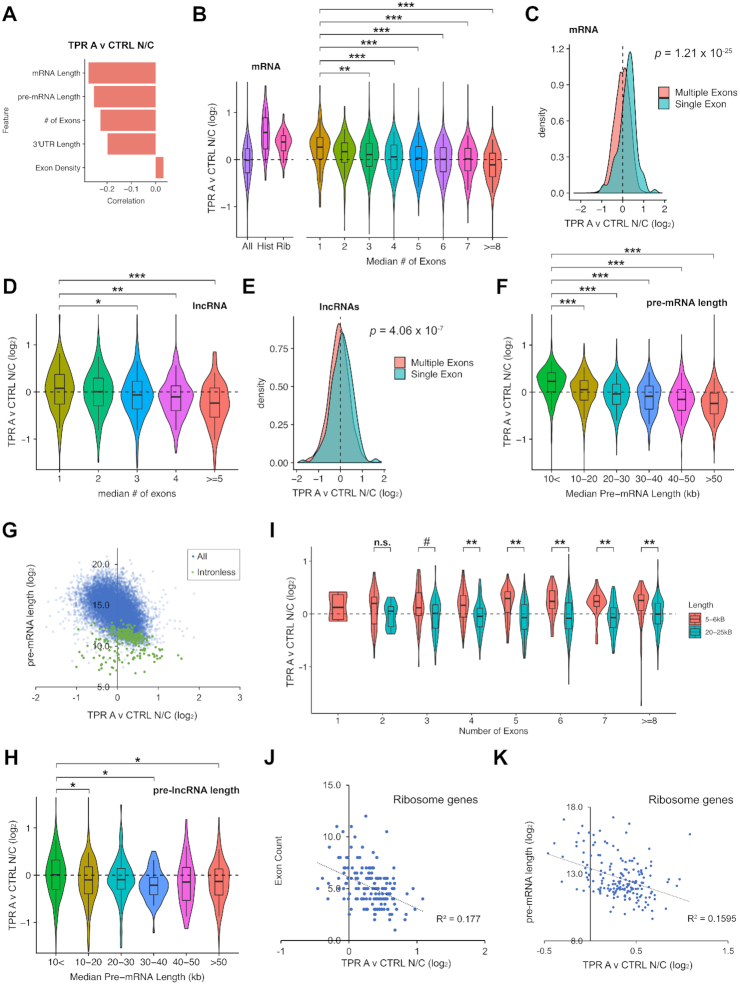
Figure 4.
TPR-depletion inhibits the nuclear export of mRNAs and lncRNAs transcribed from short pre-mRNAs, including intronless and intron-poor genes. (A) Regression analysis of TPR-dependent nuclear export (TPR A v CTRL N/C fold change) and various RNA features. Note the inverse correlation between nuclear retention upon TPR-depletion (TPR A v CTRL N/C fold change) and gene length, pre-mRNA length, number of exons and 3′UTR length. (B) Violin plots of TPR-dependent nuclear export (TPR A v CTRL N/C fold change) for various classes of mRNAs. Note that mRNAs from histone and ribosomal genes are more nuclear enriched upon TPR depletion (positive fold change value) compared to all mRNAs. Also note that the nuclear export of mRNAs generated from few exons are more sensitive to TPR-depletion than other mRNAs. (C) Density plot showing that mRNAs from intronless genes are more nuclear enriched in TPR-depleted cells compared to those with multiple exons. (D) Violin plots of TPR-dependent nuclear export (TPR A v CTRL N/C fold change) for lncRNAs with various numbers of introns. Note that the nuclear export of lncRNAs generated from few exons are more sensitive to TPR-depletion than other lncRNAs. (E) Density plot showing that lncRNAs from intronless genes are slightly more nuclear enriched in TPR-depleted cells compared to those with multiple exons. (F) Violin plots of TPR-dependent nuclear export (TPR A v CTRL N/C fold change) for pre-mRNAs of varying lengths in 10 kb windows. Again, note the inverse correlation between pre-mRNA length and nuclear retention upon TPR-depletion. (G) Scatter plot of TPR-dependent nuclear export (TPR A v CTRL N/C fold change) and pre-mRNA length. Note that the inverse correlation between nuclear retention upon TPR-depletion and pre-mRNA lengths. Intronless mRNAs (green dots) are shorter in length compared to all other pre-mRNAs (blue dots) and are most dependent on TPR-dependent nuclear export. (H) Violin plots of TPR-dependent nuclear export (TPR A v CTRL N/C fold change) for pre-lncRNAs of varying lengths in 10 kb windows. The trend for pre-lncRNAs is similar to pre-mRNAs, but again not as strong. For details, see text. (I) Violin plots of TPR-dependent nuclear export (TPR A v CTRL N/C fold change) for pre-mRNAs that are 5–6 kb or 20–25 kb in length with various numbers of exons. (J, K) For mRNAs from ribosomal protein genes, the degree of TPR-dependent nuclear export (TPR A v CTRL N/C fold change) was plotted against the number of exons (J) or pre-mRNA length (K). Note the weak correlation in each case. Mann-Whitney-Wilcoxon statistical tests with Bejamini–Hochberg adjustment for multiple comparisons was performed for B, D, F, H and I, #P > 0.05, * P < 10−3, **P < 10−5 and ***P < 10−10.