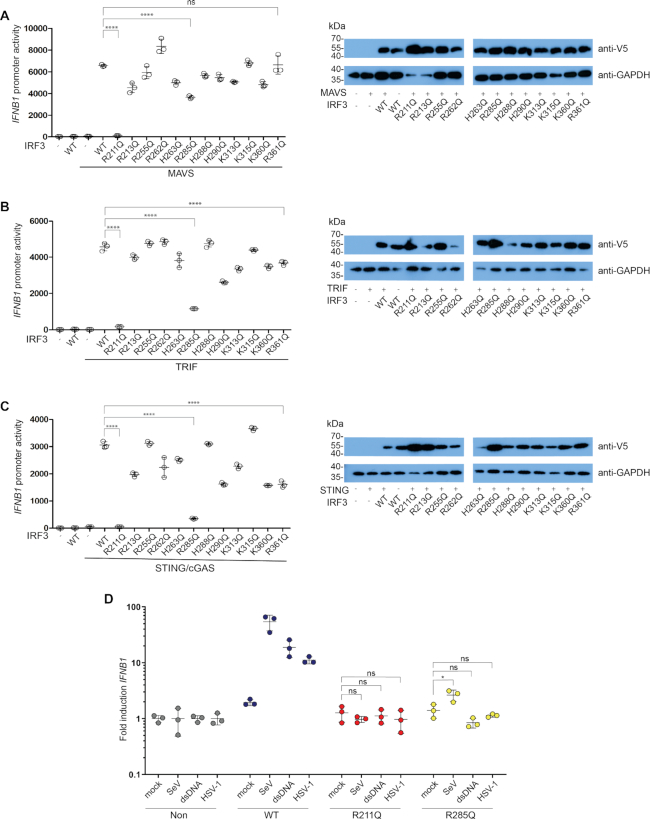
Figure 1.
Identification and validation of IRF3 mutants with impaired activation. (A–C) IRF3-deficient HEK293T cells were transiently transfected with IFNB1 promoter firefly luciferase reporter, a constitutively active Renilla luciferase reporter and either WT IRF3 or mutant IRF3 as indicated. The cells were stimulated with MAVS (A), TRIF (B) or STING and cGAS (C) and luciferase activities were measured 24 h post transfection. Firefly luciferase activity was normalized to Renilla luciferase activity and presented as triplicates ± SD. Similar data were obtained for three independent experiments (A+B) and for two independent experiments (C) (Supplementary Figure S1). (A–C) IRF3 and GAPDH amounts were detected by western blotting using an anti-V5 antibody for the detection of IRF3 and an anti-GAPDH antibody for the detection of GAPDH as a loading control. (D) IRF3-deficient THP-1 cells transduced with either IRF3 WT, IRF3 R211Q or IRF3 R285Q were infected for 6 h with SeV (2 HAU/ml) or HSV-1 (MOI 9) or stimulated for 6 h with 1 μg dsDNA. Total RNA was harvested and subjected to RT-qPCR for measurement of IFNB1. IFNB1 mRNA levels were calculated relative to GAPDH expression and normalized to the non-transduced cells for each stimulation and is shown as triplicates ± SD. Similar data were obtained for two independent experiments. (A–D) One-way ANOVA test was used for statistical analysis. ns, not significant; *, P ≤ 0.05; ****, P ≤ 0.0001.