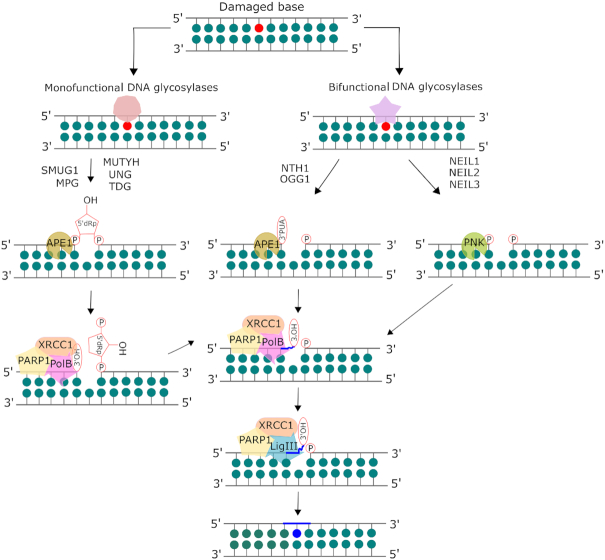
Figure 2.
Mammalian base excision repair (BER) pathway. The base lesion is excised by a lesion-specific DNA glycosylase. Monofunctional glycosylases break the glycosidic bond between the damaged base and the sugar moiety, resulting in an abasic site. AP endonuclease 1 (APE1) processes the abasic site to form a 3′OH and a deoxyribose-5′-phosphate (dRP), which is removed by the lyase activity of DNA polymerase β (pol β). Bifunctional glycosylases utilize their AP lyase activity to cleave the phosphate backbone, creating a single strand break, leaving a free 5′ phosphate and either a 3′-phospho-α, β-unsaturated aldehyde (3′-PUA) (β-elimination) or a 3′ phosphate (β,δ-elimination). APE1 acts on the β-elimination product while polynucleotide kinase phosphate (PNKP) is required to process the 3′phosphate after β,δ-elimination. The resulting 3′OH is bound by PARP1 which recruits the BER complex consisting of pol β, XRCC1 and DNA ligase. The one base gap is then filled by pol β and the nick in the DNA is sealed by DNA ligase. Adapted from Kumar et al. (8) with permission.