Key Points
Question
In patients with cancer, is the detection of pathogenic germline genetic variation improved by incorporation of automated deep learning technology compared with standard methods?
Findings
In this cross-sectional analysis of 2 retrospectively collected convenience cohorts of patients with prostate cancer and melanoma, more pathogenic variants in 118 cancer-predisposition genes were found using deep learning technology compared with a standard genetic analysis method (198 vs 182 variants identified in 1072 patients with prostate cancer; 93 vs 74 variants identified in 1295 patients with melanoma).
Meaning
The number of cancer-predisposing pathogenic variants identified in patients with prostate cancer and melanoma depends partially on the automated approach used to analyze sequence data, but further research is needed to understand possible implications for clinical management and patient outcomes.
Abstract
Importance
Less than 10% of patients with cancer have detectable pathogenic germline alterations, which may be partially due to incomplete pathogenic variant detection.
Objective
To evaluate if deep learning approaches identify more germline pathogenic variants in patients with cancer.
Design, Setting, and Participants
A cross-sectional study of a standard germline detection method and a deep learning method in 2 convenience cohorts with prostate cancer and melanoma enrolled in the US and Europe between 2010 and 2017. The final date of clinical data collection was December 2017.
Exposures
Germline variant detection using standard or deep learning methods.
Main Outcomes and Measures
The primary outcomes included pathogenic variant detection performance in 118 cancer-predisposition genes estimated as sensitivity, specificity, positive predictive value (PPV), and negative predictive value (NPV). The secondary outcomes were pathogenic variant detection performance in 59 genes deemed actionable by the American College of Medical Genetics and Genomics (ACMG) and 5197 clinically relevant mendelian genes. True sensitivity and true specificity could not be calculated due to lack of a criterion reference standard, but were estimated as the proportion of true-positive variants and true-negative variants, respectively, identified by each method in a reference variant set that consisted of all variants judged to be valid from either approach.
Results
The prostate cancer cohort included 1072 men (mean [SD] age at diagnosis, 63.7 [7.9] years; 857 [79.9%] with European ancestry) and the melanoma cohort included 1295 patients (mean [SD] age at diagnosis, 59.8 [15.6] years; 488 [37.7%] women; 1060 [81.9%] with European ancestry). The deep learning method identified more patients with pathogenic variants in cancer-predisposition genes than the standard method (prostate cancer: 198 vs 182; melanoma: 93 vs 74); sensitivity (prostate cancer: 94.7% vs 87.1% [difference, 7.6%; 95% CI, 2.2% to 13.1%]; melanoma: 74.4% vs 59.2% [difference, 15.2%; 95% CI, 3.7% to 26.7%]), specificity (prostate cancer: 64.0% vs 36.0% [difference, 28.0%; 95% CI, 1.4% to 54.6%]; melanoma: 63.4% vs 36.6% [difference, 26.8%; 95% CI, 17.6% to 35.9%]), PPV (prostate cancer: 95.7% vs 91.9% [difference, 3.8%; 95% CI, –1.0% to 8.4%]; melanoma: 54.4% vs 35.4% [difference, 19.0%; 95% CI, 9.1% to 28.9%]), and NPV (prostate cancer: 59.3% vs 25.0% [difference, 34.3%; 95% CI, 10.9% to 57.6%]; melanoma: 80.8% vs 60.5% [difference, 20.3%; 95% CI, 10.0% to 30.7%]). For the ACMG genes, the sensitivity of the 2 methods was not significantly different in the prostate cancer cohort (94.9% vs 90.6% [difference, 4.3%; 95% CI, –2.3% to 10.9%]), but the deep learning method had a higher sensitivity in the melanoma cohort (71.6% vs 53.7% [difference, 17.9%; 95% CI, 1.82% to 34.0%]). The deep learning method had higher sensitivity in the mendelian genes (prostate cancer: 99.7% vs 95.1% [difference, 4.6%; 95% CI, 3.0% to 6.3%]; melanoma: 91.7% vs 86.2% [difference, 5.5%; 95% CI, 2.2% to 8.8%]).
Conclusions and Relevance
Among a convenience sample of 2 independent cohorts of patients with prostate cancer and melanoma, germline genetic testing using deep learning, compared with the current standard genetic testing method, was associated with higher sensitivity and specificity for detection of pathogenic variants. Further research is needed to understand the relevance of these findings with regard to clinical outcomes.
This study compares pathogenic variant detection accuracy in cancer-disposition genes of prostate cancer and melanoma cohort biopsy samples using deep learning vs standard genetic analysis methods.
Introduction
Germline genetic testing is increasingly used to identify a class of inherited genetic changes called pathogenic variants, which are associated with an increased risk of developing cancer and other diseases. The detection of pathogenic variants associated with cancer can identify patients and families with inherited cancer susceptibility in whom established gene-specific screening recommendations can be implemented.1,2 Furthermore, germline genetic testing of patients with cancer can identify pathogenic variant carriers who tend to have genetically determined greater response to chemotherapy and targeted anticancer agents.3,4 However, even when the clinical presentation is highly suggestive of a particular genetic cancer-predisposition syndrome, only a small fraction of patients are found to carry germline pathogenic variants,5,6,7,8 raising concern about the possibility of incomplete detection of known or expected pathogenic variants by the current standard germline variant detection methods, which includes the Genome Analysis Toolkit.9
Computational methods that use deep learning neural networks, which incorporate layers of networks to learn and analyze complex patterns in data, have demonstrated superior performance compared with standard methods for disease recognition,10 pathological and radiological image analysis,11 and natural language processing.12 Deep learning methods also have shown enhanced germline variant detection compared with standard methods in laboratory samples with known genetic variation.13 However, it is unknown whether using deep learning approaches can result in identifying additional patients with pathogenic variants missed by the current standard analysis framework. In this study, it was hypothesized that deep learning variant detection would have higher sensitivity compared with the standard method for identifying clinically relevant pathogenic variants when applied to clinical samples from patients with cancer.
Methods
Ethics Approval and Consent to Participate
Written informed consent from patients and institutional review board approval, allowing comprehensive genetic analysis of germline samples, were obtained by the original studies that enrolled patients in the US and Europe between 2010 and 2017. The final date of clinical data collection was December 2017 (eMethods in Supplement 1). The secondary genomic and deep learning analyses performed for this study were approved under Dana-Farber Cancer Institute institutional review board protocols 19-139 and 02-293. This study conforms to the Declaration of Helsinki.
Patient Cohorts and Genomic Data Collection
Publicly available germline whole-exome sequencing data of 2 independent sets of published cohorts, each of which comprised convenience samples, were included in this study (Figure 1). One cohort of patients with prostate cancer was described by Armenia et al,14 and a second cohort of patients with melanoma was obtained from 10 publicly available data sets (eMethods in Supplement 1). All germline whole-exome sequencing data were generated by the original studies using paired-end, short-read Illumina platforms (Illumina Inc). Patient cohorts were not selected for a positive family history of cancer or early-onset disease. Germline genetic data of all cohorts were available for analysis. This analysis was not intended to change the management of the study cohorts.
Figure 1. Overview of the Study Design and the Germline Variant Detection Methods Used.
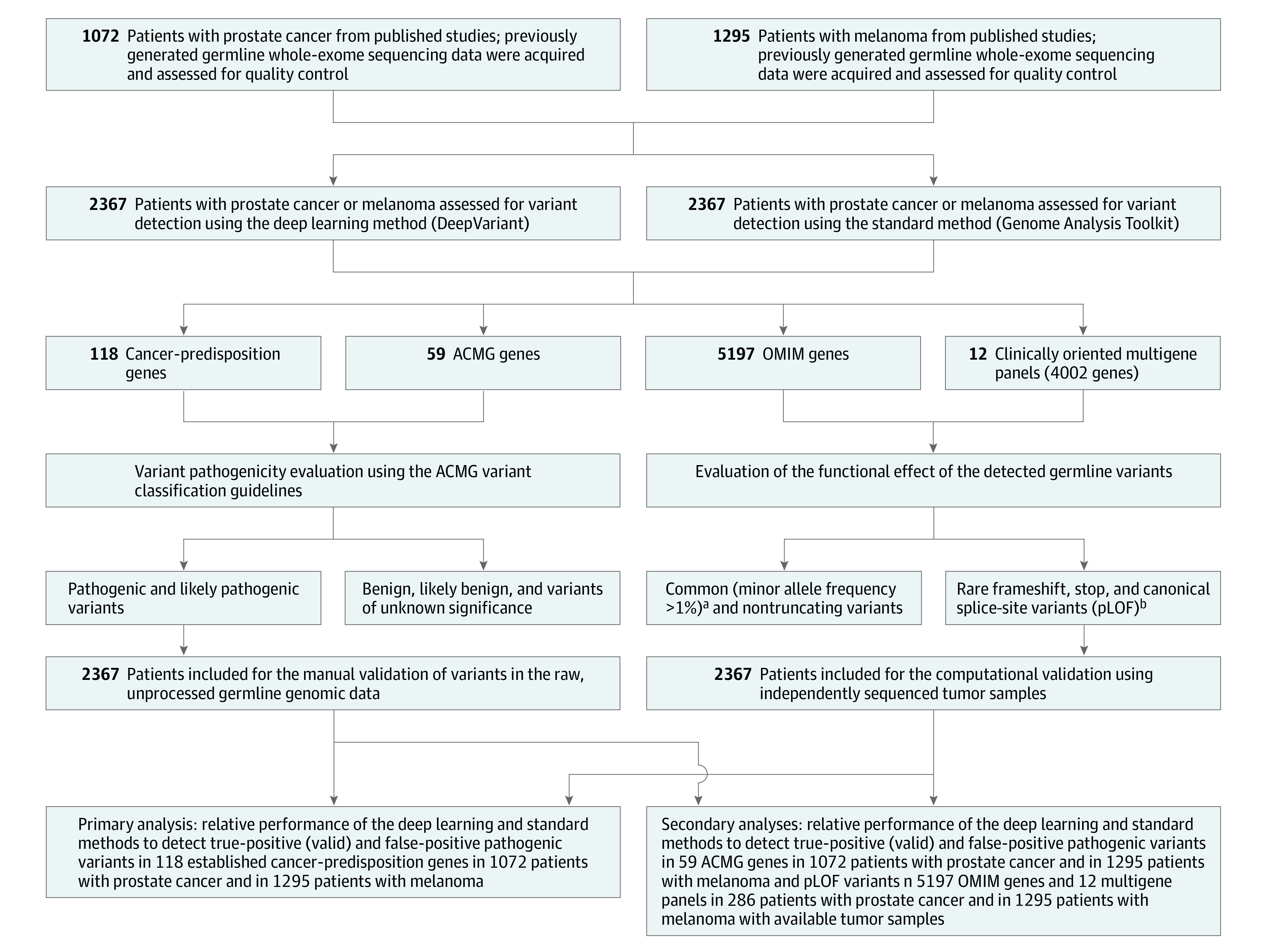
Germline whole-exome sequencing data of 2 independent cohorts were analyzed using a deep learning variant detection approach and the current standard germline variant detection method to assess the sensitivity, specificity, positive predictive value, and negative predictive value of detecting pathogenic variants in 118 cancer-predisposition genes as well as in 59 American College of Medical Genetics and Genomics (ACMG) genes. In addition, an exome-wide analysis of putative loss-of-function (pLOF) variants in 5197 Online Mendelian Inheritance in Man (OMIM) genes and in 12 clinically oriented multigene panels was conducted in 286 patients with prostate cancer and in 1295 patients with melanoma whose tumors were available for independent variant validation. For each analysis, the performance of the deep learning method and the standard method was compared with the performance obtained when both methods are concurrently used (additional information appears in the Methods section). The performance of the standard and deep learning methods to detect pathogenic variants in the cancer-predisposition and ACMG gene sets was tested on all patients with prostate cancer and melanoma (n = 2367). However, the performance of the 2 methods in the OMIM gene set and the multigene panels was tested on 286 patients with prostate cancer and 1295 patients with melanoma (n = 1581) whose tumor sequencing data were available for computational validation of the identified pLOF variants.
aRepresents the frequency of the allele (also called variant) in the general population.
bGenetic variants that are likely to severely disrupt the protein function by truncating the gene transcript. Examples of pLOF variants include frameshift, stop-codon, and canonical splice-site variants.
Germline Variant Detection Methods
The Genome Analysis Toolkit (version 3.7),15 the most widely used germline variant detection method,16,17,18 was considered the standard method in this analysis and the DeepVariant method (version 0.6.0) was used to perform deep learning variant detection13,19 (Figure 1 and eFigure 1 in Supplement 1). The standard and deep learning methods were run using the recommended parameters.20,21 Details of the standard and deep learning analysis frameworks (along with the corresponding computer program codes) appear in the eNotes in Supplement 1.
Selection of Mendelian Gene Sets
In this study, pathogenic variants in 118 established cancer-predisposition genes and 59 mendelian high-penetrance genes deemed clinically actionable by the American College of Medical Genetics and Genomics (ACMG; collectively called the ACMG gene set) were analyzed (eTable 1 in Supplement 2). Patients with cancer can also be carriers of disease-causing variants in autosomal recessive and low-penetrant genes unrelated to cancer, leading to a gene product that has decreased or no function (termed putative loss of function; pLOF). Thus, pLOF variants in 5197 clinically relevant genes in the Online Mendelian Inheritance in Man (OMIM) database (collectively called the OMIM gene set) (eTable 1 in Supplement 2) and 12 clinically oriented multigene panels (eMethods in Supplement 1 and eTable 2 in Supplement 3) also were characterized.
Germline Variant Pathogenicity Evaluation
The identified germline variants in the cancer-predisposition gene set and in the ACMG gene set were independently classified by 2 clinical geneticists (S.H.A. and L.W.) into the 5 categories of benign, likely benign, variants of unknown significance, likely pathogenic, and pathogenic using established ACMG guidelines.22 Only pathogenic and likely pathogenic variants were included in this study (hereafter collectively referred to as pathogenic variants).
Validation of Identified Germline Variants
Given the lack of a criterion reference standard to independently validate the results of the standard genetic analysis method and the deep learning method at every position of each gene in each participant in this study, an established manual validation framework was adopted.23 In this framework, variants in the cancer-predisposition and ACMG gene sets were manually evaluated in an independent blind fashion by 3 examiners (S.H.A., J.R.C., and A.T.-W.) using a genetic data visualization tool called the Integrative Genomics Viewer.24 Identified pathogenic variants were judged to be valid (ie, true-positive) if they were deemed present in the raw genomic data by at least 2 of the 3 examiners. Otherwise, the variant was judged to be false-positive (eTable 3 in Supplement 1). Germline pLOF variants in the OMIM gene set and in the multigene panel analyses were judged to be valid by examining the independently sequenced tumor samples from the same patient for the presence of these variants (eMethods in Supplement 1). Genomic regions in which no pathogenic variants were identified by either method were not manually validated for the absence of variants and were assumed to be devoid of pathogenic variation.
Calculating the Performance of the 2 Methods
Because of the lack of an independently generated criterion reference standard variant set for the analyzed data, the set of variants generated by combining all variants judged to be valid from either the standard genetic analysis approach or the deep learning method was used as a reference to evaluate each method’s performance (hereafter referred to as the reference variant set). Consequently, true sensitivity and specificity could not be calculated due to the lack of a criterion reference standard. Therefore, the terms sensitivity and specificity were defined and calculated for method comparisons. Sensitivity was defined as the proportion of true-positive variants (ie, judged to be valid) identified by each method to the total number of true-positive variants in the reference variant set. Specificity was defined as the proportion of true-negative variants determined by each method to the total number of true-negative variants in the reference variant set (eTable 3 and eMethods in Supplement 1).
In addition, given the rare nature of disease-causing pathogenic variants, and to enhance study power, the sensitivity, specificity, positive predictive value (PPV), and negative predictive value (NPV) for the standard method and the deep learning method were calculated using 3 predefined gene sets (cancer predisposition, ACMG, and OMIM). In this study, PPV and NPV solely referred to the probability that a participant with a variant identified by these methods had (or did not have) the molecular genetic variant, not the clinical disease phenotype. Detailed definitions of the performance metrics used in this study appear in eTable 3 in Supplement 1. In addition, the performance of using the standard method and the deep learning method in tandem (combined methods) was evaluated by assessing the number of pathogenic and pLOF variants identified by either the standard method or the deep learning method and judged to be valid.
Outcomes
The primary outcomes were defined as the absolute number of identified pathogenic variants judged to be valid on manual review and the sensitivity, specificity, PPV, and NPV of the deep learning and standard methods in the cancer-predisposition gene set (Figure 1). Secondary outcomes included the absolute number of pathogenic and pLOF variants judged to be valid and the sensitivity, specificity, PPV, and NPV of each method using the ACMG, OMIM, and multigene panel sets (Figure 1).
Statistical Analysis
Two-sided χ2 tests were used to calculate the P values and 95% CIs for the differences in sensitivity, specificity, PPV, and NPV for each method. In addition, 2-sided binomial tests were used to calculate the 95% CIs for the proportions. Two-sided Mann-Whitney tests were used to evaluate the differences in the sequencing depth of the examined genomic regions. For the analysis of the clinical characteristics in the examined cohorts, patients with clinical data not reported in the originating study were included in a “not reported” category for each clinical characteristic.
To evaluate the performance of the standard and deep learning methods, pathogenic variants in the cancer-predisposition and ACMG gene sets (total: 151 genes) were combined to calculate the receiver operating characteristic curve by using quality score thresholds for variants as follows. For each threshold, we used the standard and deep learning models to determine if the assessed variant was real or artifactual. These decisions were then compared with the results of the manual validation of these pathogenic variants and the true-positive, true-negative, false-positive, and false-negative rates were calculated (eTable 3 and eMethods in Supplement 1). An assumption was made that positions in the genome with no variants identified by the standard or deep learning approach were true-negative variants.
To evaluate the computational effects of defining true-positive variants as judged to be valid variants by all 3 examiners for the primary outcomes of this study, a post hoc analysis was conducted comparing the absolute number and the fraction of the manually validated pathogenic variants in the cancer-predisposition gene set identified by each method vs the total number of variants identified by each method using 2-sided χ2 tests. P values <.05 were considered statistically significant without adjustment for multiple comparisons. The findings for the secondary analyses should be interpreted as exploratory. Statistical analyses were performed using the exact2x2 (version 1.5.2), binom (version 1.1.1), and stats (version 3.5.1) packages on R version 3.5.1 (R Foundation for Statistical Computing).
Results
Sequencing Metrics and Overall Germline Variant Detection
The mean age of diagnosis was 63.7 years (SD, 7.9 years) for the prostate cancer cohort (n = 1072 men) and was 59.8 years (SD, 15.6 years) for the melanoma cohort (n = 1295; 488 [37.7%] women) (Table 1). Germline variants in the prostate cancer and melanoma cohorts were analyzed using the standard and deep learning methods (Figure 1 and eFigure 1 in Supplement 1). The mean exome-wide sequencing depth was 105.78 reads (SD, 52.92 reads) for the prostate cancer cohort and was 86.85 reads (SD, 45.27 reads) for the melanoma cohort (eFigure 2 in Supplement 1). Of 37 373 535 germline genetic variants identified in 1072 germline prostate cancer exomes, 92.1% were identified by both the standard and deep learning methods (eFigure 3 in Supplement 1), demonstrating a discrepancy between these variant detection approaches.
Table 1. Characteristics of the Study Cohorts.
| Clinical characteristic | Prostate cohort (n = 1072) | Melanoma cohort (n = 1295) |
|---|---|---|
| Age at diagnosis, mean (SD), y | 63.7 (7.9) | 59.8 (15.6) |
| Age not reported in original study, No. (%) | 352 (32.8) | 115 (8.9) |
| Sex, No. (%) | ||
| Female | 0 | 488 (37.7) |
| Male | 1072 (100.0) | 752 (58.1) |
| Sex not reported in original study, No. (%) | 0 | 55 (4.2) |
| Disease status, No. (%) | ||
| Primary | 467 (43.6) | 278 (21.5) |
| Metastatic | 590 (55.0) | 990 (76.4) |
| Disease status not reported in original study, No. (%) | 15 (1.4) | 27 (2.1) |
| Genetic ancestry, No. (%)a | ||
| European | 857 (79.9) | 1060 (81.9) |
| South Asianb | 80 (7.5) | 157 (12.1) |
| African American | 78 (7.3) | 4 (0.3) |
| Admixed Americanc | 40 (3.7) | 60 (4.6) |
| East Asiand | 16 (1.5) | 14 (1.1) |
| Unknown | 1 (0.1) | 0 |
Inferred using genetic variance and genetic similarity and by using samples from the 1000 Genome as reference samples. A more detailed description of this analysis appears in the eMethods in Supplement 1.
Included individuals from Bangladesh, Pakistan, and India.
Included individuals with Mexican ancestry from the US, Puerto Rico, Colombia, and Peru.
Included individuals from China, Japan, and Vietnam.
Primary Analysis
Detection of Pathogenic Variants in the Cancer-Predisposition Gene Set
A total of 171 pathogenic variants were identified by both the standard and deep learning methods in 118 cancer-predisposition genes in the prostate cancer cohort (n = 1072). The deep learning method exclusively identified 36 pathogenic variants, of which 27 (75% [95% CI, 58.9% to 86.2%]) were judged to be valid (true-positive findings) and 9 (25% [95% CI, 13.8% to 41.1%]) were judged to be false-positive findings (Figure 2A, Table 2, and eTable 4 in Supplement 1). The standard method exclusively identified 27 pathogenic variants, of which 11 (40.7% [95% CI, 24.5% to 59.3%]) were judged to be valid (true-positive findings) and 16 (59.3% [95% CI, 40.7% to 75.5%]) were judged to be false-positive findings (Figure 2B, Table 2, and eTable 5 in Supplement 1).
Figure 2. Examples of Germline Pathogenic Variants That Were Exclusively Identified by the Deep Learning or Standard Variant Detection Methods.
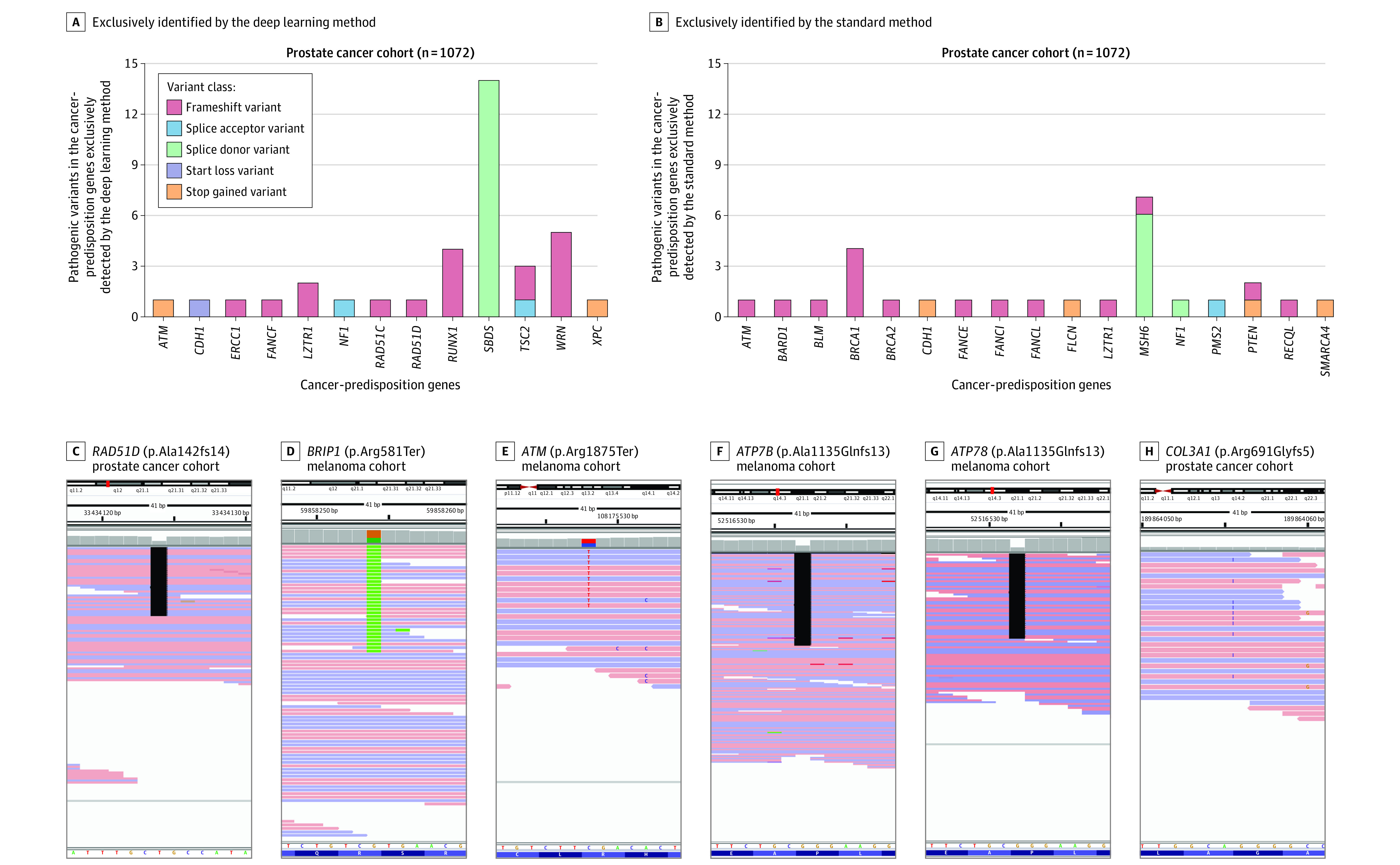
Thirty-six germline pathogenic variants were identified by the deep learning method, 75% of which were judged to be valid or true-positive (A), whereas 27 variants were identified by the standard method, 40.7% of which were judged to be valid (B). A similar analysis of 1295 patients with melanoma showed a similarly higher detection rate with the deep learning method (eFigure 4). Examples show pathogenic variants in the cancer-predisposition gene set (C-E) and the clinically actionable American College of Medical Genetics and Genomics gene set (F-H) that were only identified by the deep learning method and judged to be valid on manual assessment.
Table 2. Performance of the Deep Learning and Standard Germline Variant Detection Methods.
| Tested gene seta | Identified variants, No. | No. of putatively true-positive variants identified by both methods | No. of putatively true-positive variants and false-positive identified by each method | Putatively true-positive variants | Putatively false-positive variants | Sensitivity (95% CI)b | Specificity (95% CI)b | PPV (95% CI)c | NPV (95% CI)c | |||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Pathogenic or pLOF | Putatively true-positive | No. identified by each method | % of all variants | No. identified by each method | % of all variants | |||||||
| Cancer predisposition (118 genes) | ||||||||||||
| Prostate cancer (n = 1072) | ||||||||||||
| Deep learning method | 207 | 198 | 171 | 36 | 27 | 95.7 | 9 | 4.3 | 94.7 (90.8-97.0) | 64.0 (44.5-79.8) | 95.7 (91.9-97.7) | 59.3 (40.7-75.5) |
| Standard method | 198 | 182 | 171 | 27 | 11 | 91.9 | 16 | 8.1 | 87.1 (81.9-91.0) | 36.0 (20.2-55.5) | 91.9 (87.3-95.0) | 25.0 (13.8-41.1) |
| Combined methods | 234 | 209 | 89.3 | 25 | 10.7 | |||||||
| Melanoma (n = 1295) | ||||||||||||
| Deep learning method | 171 | 93 | 42 | 129 | 51 | 54.4 | 78 | 45.6 | 74.4 (66.1-81.2) | 63.4 (56.7-69.6) | 54.4 (46.9-61.7) | 80.8 (74.2-86.1) |
| Standard method | 209 | 74 | 42 | 167 | 32 | 35.4 | 135 | 64.6 | 59.2 (50.4-67.4) | 36.6 (30.4-43.3) | 35.4 (29.2-42.1) | 60.5 (51.8-68.5) |
| Combined methods | 338 | 125 | 37.0 | 213 | 63.0 | |||||||
| American College of Medical Genetics and Genomics (ACMG; 59 genes) | ||||||||||||
| Prostate cancer (n = 1072) | ||||||||||||
| Deep learning method | 121 | 111 | 100 | 21 | 11 | 91.7 | 10 | 8.3 | 94.9 (89.3-97.6) | 64.3 (45.8-79.3) | 91.7 (85.5-95.4) | 75.0 (55.1-88.0) |
| Standard method | 124 | 106 | 100 | 24 | 6 | 85.5 | 18 | 14.5 | 90.6 (83.9-94.7) | 35.7 (20.7-54.2) | 85.5 (78.2-90.6) | 47.6 (28.3-67.6) |
| Combined methods | 145 | 117 | 80.7 | 28 | 19.3 | |||||||
| Melanoma (n = 1295) | ||||||||||||
| Deep learning method | 115 | 48 | 17 | 98 | 31 | 41.7 | 67 | 58.3 | 71.6 (59.9-81.0) | 49.2 (40.9-57.7) | 41.7 (33.1-50.9) | 77.4 (67.4-85.0) |
| Standard method | 101 | 36 | 17 | 84 | 19 | 35.6 | 65 | 64.4 | 53.7 (41.9-65.1) | 50.8 (42.3-59.1) | 35.6 (27.0-45.4) | 68.4 (58.6-76.7) |
| Combined methods | 199 | 67 | 33.7 | 132 | 66.3 | |||||||
| Online Mendelian Inheritance in Man (OMIM; 5197 genes) | ||||||||||||
| Prostate cancer (n = 286)d | ||||||||||||
| Deep learning method | 753 | 708 | 673 | 80 | 35 | 94.0 | 45 | 6.0 | 99.7 (99.0-99.9) | 11.8 (5.5-23.4) | 94.0 (92.1-95.5) | 75.0 (40.9-92.9) |
| Standard method | 681 | 675 | 673 | 8 | 2 | 99.1 | 6 | 0.9 | 95.1 (93.2-96.4) | 88.2 (76.6-94.5) | 99.1 (98.1-99.6) | 56.3 (45.3-66.6) |
| Combined methods | 761 | 710 | 93.3 | 51 | 6.7 | |||||||
| Melanoma (n = 1295) | ||||||||||||
| Deep learning method | 1042 | 619 | 526 | 516 | 93 | 59.4 | 423 | 40.6 | 91.7 (89.4-93.6) | 30.8 (27.2-34.5) | 59.4 (56.4-62.3) | 77.0 (71.4-81.9) |
| Standard method | 770 | 582 | 526 | 244 | 56 | 75.6 | 188 | 24.4 | 86.2 (83.4-88.6) | 69.2 (65.5-72.8) | 75.6 (72.4-78.5) | 82.0 (78.4-85.0) |
| Combined methods | 1286 | 675 | 52.5 | 611 | 47.5 | |||||||
Abbreviations: NPV, negative predictive value; pLOF, putative loss-of-function; PPV, positive predictive value.
The performance of the deep learning and standard methods to correctly detect pathogenic and pLOF variants was assessed in 2 convenience cohorts of patients with prostate cancer or melanoma. Given the lack of a criterion reference standard to independently validate the findings of the deep learning and standard methods, each method’s performance (ie, sensitivity, specificity, PPV, and NPV) was compared with the performance obtained by concurrently running both methods (combined methods) on the analyzed gene set (additional details appear in the Methods section).
Sensitivity was defined as the proportion of true-positive (valid) variants identified by each method to the total number of true-positive variants in the combined variant set (ie, the reference variant set). Specificity was defined as the proportion of true-negative variants determined by each method to the total number of true-negative variants in the reference variant set. Considering the lack of an independently generated criterion reference standard variant set for the analyzed data, the results derived from the combined methods were considered the reference variant set that was used to calculate each method’s performance.
Prostate cancer and melanoma patient cohorts were used to calculate the PPV and the NPV in whom germline analysis of the cancer predisposition genes are frequently done. However, these patient cohorts are not commonly tested for the noncancer ACMG or OMIM gene sets, so the calculated PPV and NPV for these gene sets may not represent the actual PPV and NPV of the standard and deep learning methods for patients in whom testing using these gene sets is indicated. Furthermore, the majority of patients included in this study were of European ancestry, so the performance of these methods in patient cohorts of other backgrounds may differ.
There were fewer patients with available tumor sequencing data for independent validation.
In the prostate cancer cohort, the deep learning method had higher sensitivity compared with the standard method (94.7% vs 87.1%, respectively; difference, 7.6% [95% CI, 2.2% to 13.1%]; P = .006), higher specificity (64.0% vs 36.0%; difference, 28.0% [95% CI, 1.4% to 54.6%]; P = .047), and higher NPV (59.3% vs 25.0%; difference, 34.3% [95% CI, 10.9% to 57.6%]; P = .006) (Table 2). However, the PPV for the deep learning method was not significantly different from the standard method (95.7% vs 91.9%, respectively; difference, 3.8% [95% CI, –1.0% to 8.4%]; P = .11).
The pathogenic variants exclusively identified by the deep learning method in the cancer-predisposition genes were more likely to be judged valid on manual review compared with variants exclusively identified by the standard method (75.0% vs 40.7%, respectively; difference, 34.3% [95% CI, 10.9% to 57.6%]; P = .006). Overall, the deep learning method identified 16 more patients with prostate cancer who had pathogenic variants associated with elevated cancer risk that were missed by the standard method and were judged to be valid on manual review (Table 2).
To explore the generalizability of these findings, germline whole-exome sequencing data from 1295 patients with melanoma also were analyzed. The deep learning method identified more patients with pathogenic variants judged to be valid (true-positive findings) compared with the standard method (93 vs 74, respectively) and identified fewer pathogenic variants judged to be false-positive findings (78 vs 135) (Table 2 and eFigure 4 and eTables 6 and 7 in Supplement 1). The deep learning method had higher sensitivity compared with the standard method (74.4% vs 59.2%, respectively; difference, 15.2% [95% CI, 3.7% to 26.7%]; P = .01), higher specificity (63.4% vs 36.6%; difference, 26.8% [95% CI, 17.6% to 35.9%]; P < .001), higher PPV (54.4% vs 35.4%; difference, 19.0% [95% CI, 9.1% to 28.9%]; P < .001), and higher NPV (80.8% vs 60.5%; difference, 20.3% [95% CI, 10.0% to 30.7%]; P < .001) (Table 2). Furthermore, pathogenic variants in the cancer-predisposition genes exclusively identified by the deep learning method were significantly more likely to be judged valid on manual review compared with those exclusively identified by the standard method (39.5% vs 19.2%, respectively; difference, 20.3% [95% CI, 10.0% to 30.7%]; P < .001).
The use of both the deep learning and standard methods in tandem (combined methods) resulted in the highest number of manually validated pathogenic variants in the cancer-predisposition genes. In the prostate cohort, there were 182 pathogenic variants judged to be valid (true-positive) using the standard method, 198 variants using the deep learning method, and 209 variants using combined methods. In the melanoma cohort, there were 74 pathogenic variants judged to be valid using the standard method, 93 variants using the deep learning method, and 125 variants using combined methods (Table 2).
Pathogenic variants exclusively identified by the deep learning method and judged to be valid on manual review included a frameshift in RAD51D (OMIM: 602954) (p.Ala142GlnfsTer14; rs730881935) that is associated with a 6-fold increased risk of ovarian cancer25 (Figure 2C), a nonsense variant in BRIP1 (OMIM: 605882) (p.Arg581Ter; rs780020495) that is associated with a 14-fold increased risk for high-grade serous ovarian cancer26 (Figure 2D), a truncating variant in ATM (OMIM: 607585) (p.Arg1875Ter; rs376603775) that, in the heterozygous state, confers a 2 to 5 times higher risk for breast, colorectal, and gastric cancers7,27 (Figure 2E), and a stop-codon variant in SDHA (OMIM: 600857) (p.Arg512Ter; rs748089700) that is associated with a 40% chance of developing pheochromocytoma or paraganglioma by 40 years of age28 (eFigure 5 in Supplement 1). The standard method also exclusively identified several clinically relevant pathogenic variants judged to be valid, including a frameshift in ATM (OMIM: 607585) (p.Thr2333AsnfsTer6; rs587781299) and several splice donor variants in MSH6 (OMIM: 600678) (NM_000179.3:c.4001 + 2del) (eTables 5 and 6 in Supplement 1).
Although the use of a more stringent criterion (agreement of 3 examiners instead of 2; eMethods in Supplement 1) reduced the absolute number and fraction of valid (true-positive) pathogenic variants identified by each approach, the deep learning method still identified significantly more true-positive pathogenic variants than the standard method regardless of the criteria or cohorts used (62.5% vs 20%, respectively, for the prostate cancer cohort, P < .001 and 23.1% vs 8.48% for the melanoma cohort, P < .001; eFigure 6 in Supplement 1).
Secondary Analyses
Detection of Pathogenic Variants in ACMG Genes
When examining the 59 ACMG genes in 1072 patients with prostate cancer, the deep learning method identified more patients with pathogenic variants judged to be valid (true-positive) on manual review than the standard method (111 vs 106, respectively) and identified fewer variants judged to be false-positive findings (10 vs 18) (eTables 8 and 9 in Supplement 1). The deep learning method achieved a higher specificity than the standard method (64.3% vs 35.7%, respectively; difference, 28.6% [95% CI, 3.5% to 53.7%]; P = .03). However, the sensitivity of the deep learning method was not significantly different than the standard method (94.9% vs 90.6%, respectively; difference, 4.3% [95% CI, –2.3% to 10.9%]; P = .21), nor was the PPV (91.7% vs 85.5%; difference, 6.2% [95% CI, –1.7% to 14.1%]; P = .12) or the NPV (75.0% vs 47.6%; difference, 27.4% [95% CI, –0.12% to 54.9%]; P = .06) (Table 2).
A similar analysis of the melanoma cohort (n = 1295) revealed that the deep learning method identified 12 more patients with clinically actionable pathogenic variants judged to be valid in the ACMG gene set compared with the standard method (48 vs 36, respectively) (eTables 10 and 11 in Supplement 1). The deep learning method yielded a higher sensitivity than the standard method (71.6% vs 53.7%, respectively; difference, 17.9% [95% CI, 1.82% to 34.0%]; P = .03). However, the specificity of the deep learning method was not significantly different than the standard method (49.2% vs 50.8%, respectively; difference, –1.6% [95% CI, –13.6% to 10.5%]; P = .81), nor was the PPV (41.7% vs 35.6%; difference, 6.1% [95% CI, –6.9% to 19.1%]; P = .36) or the NPV (77.4% vs 68.4%; difference, 9.0% [95% CI, –3.8% to 21.9%]; P = .17) (Table 2).
The use of both the deep learning and standard methods in tandem (combined methods) achieved a higher detection rate than using either of these methods independently in the ACMG gene set. In the prostate cohort, there were 106 pathogenic variants judged to be valid using the standard method, 111 using the deep learning method, and 117 using the combined methods. In the melanoma cohort, there were 36 pathogenic variants judged to be valid using the standard method, 48 using the deep learning method, and 67 using the combined methods (Table 2).
The pathogenic variants exclusively identified by the deep learning method in the ACMG genes and judged to be valid on manual review included truncating pathogenic variants in ATP7B (OMIM: 606882) (Figure 2F and 2G), a gene associated with Wilson disease and fatal liver failure, and COL3A1 (OMIM: 120180) (Figure 2H), a gene associated with autosomal dominant vascular Ehlers-Danlos syndrome complicated by early-onset aortic dissection, viscus rupture, and premature mortality. The standard method exclusively identified pathogenic variants in the ACMG genes that were judged to be valid, including pathogenic variants in SCN5A (OMIM: 600163) that are associated with cardiac arrhythmias and variants in OTC (OMIM: 300461) associated with severe metabolic hyperammonemia.
Evaluation of Model Performance
In the analyses that combined pathogenic variants in the cancer-predisposition and ACMG gene sets, the deep learning method had an area under the curve value of 0.94 (95% CI, 0.91 to 0.97) compared with 0.89 (95% CI, 0.84 to 0.93) for the standard method in the prostate cancer cohort and of 0.76 (95% CI, 0.71 to 0.81) for the deep learning method compared with 0.60 (95% CI, 0.55 to 0.67) for the standard method in the melanoma cohort (Figure 3 and eFigures 7 and 8 in Supplement 1). Results of other model performance metrics appear in eTable 12 in Supplement 1.
Figure 3. Evaluation of Model Performance and Normalized Confusion Matrices of the Standard and Deep Learning Methods.
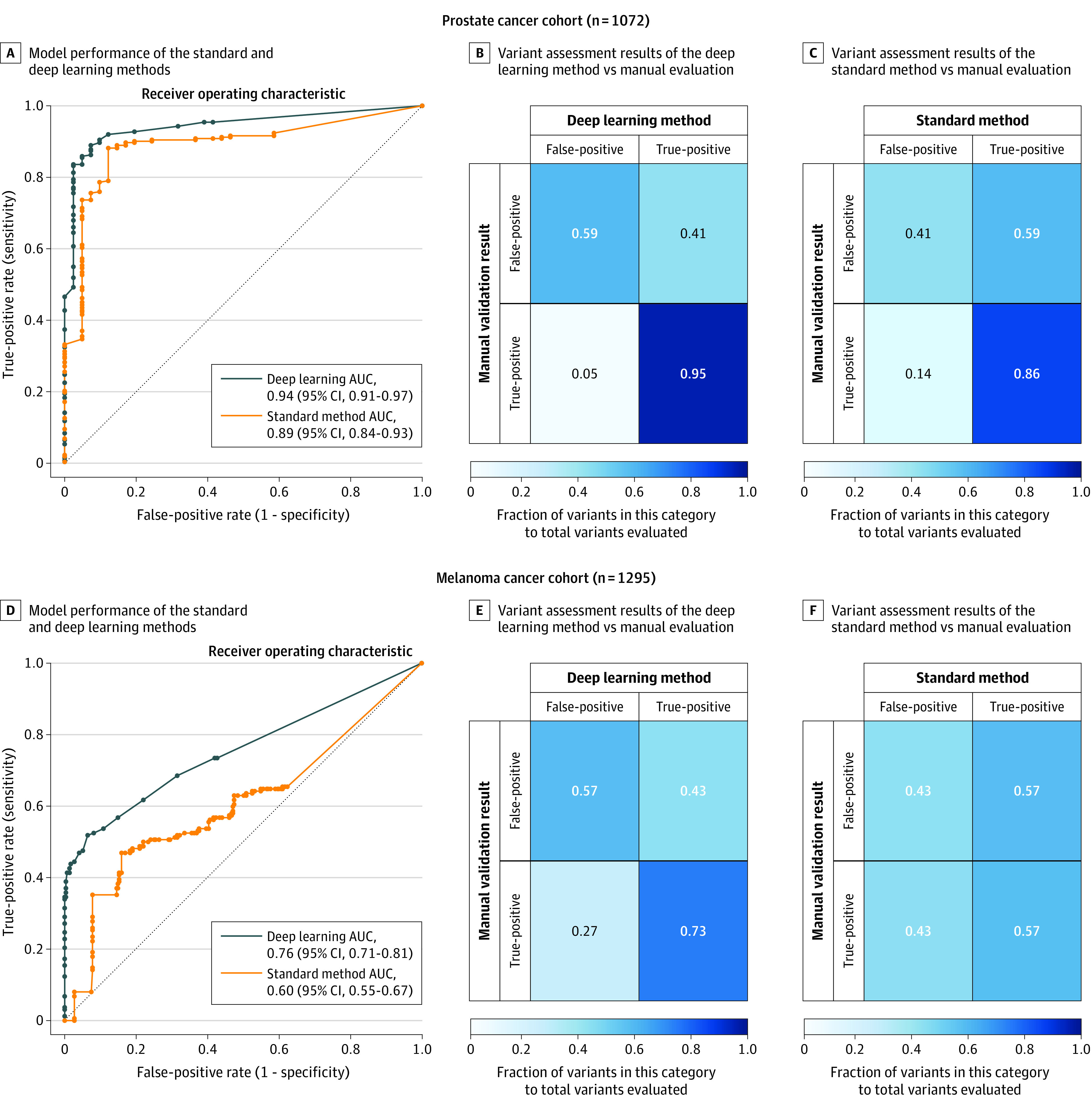
Manually assessed pathogenic variants in 151 cancer-predisposition and American College of Medical Genetics and Genomics genes were used to calculate the receiver operating characteristic curve using a set of thresholds for the quality scores. For each quality threshold, the standard and deep learning models predicted if the variant being assessed is real or artifactual. These predictions were then compared with the results of the manual validation of these pathogenic variants (judged to be valid or not) and true-positive, true-negative, false-positive, and false-negative rates were calculated. Analysis of prostate cancer cohort (n = 1072) (A). Normalized confusion matrices (B and C), representing the fractions of the manually validated true-positive and false-positive variants (ie, the reference variant set; y-axis) that were correctly identified as such by the standard method and the deep learning method (x-axis). For each matrix, the left-upper and right-lower squares represent the degree of agreement while the right-upper and left-lower squares represent the degree of mismatching between the results of manual validation and the method-based variant assessment. The intensity of the square shading represents the metric within the square, which is the ratio of the number of variants in each category to the total number of true-positive or true-negative in the reference variant set (ie, the manual validation set). Compared with the standard method, there was more agreement between the variant assessment results of the deep learning method and the manual validation results in the prostate cancer cohort (n = 1072). Analysis of the melanoma cohort (n = 1295) (D). Similarly, there was more agreement between the deep learning–based variant assessment and the manual evaluation of these variants in the raw genomic data of the melanoma cohort (E and F).
Detection of pLOF Variants in 5197 Clinically Relevant Mendelian Genes
Among 286 patients with prostate cancer whose tumors were available for independent validation, more germline pLOF variants validated in the tumor sequencing data were identified by the deep learning method than by the standard method (708 vs 675, respectively), resulting in higher sensitivity (99.7% vs 95.1%; difference, 4.6% [95% CI, 3.0% to 6.3%]; P < .001), lower specificity (11.8% vs 88.2%; difference, –76.4% [95% CI, −89.0% to −64.0%]; P < .001), and lower PPV (94.0% vs 99.1%; difference, –5.1% [95% CI, −6.9% to −3.3%]; P < .001) (Table 2). The NPV for the deep learning method was not significantly different from the standard method (75.0% vs 56.3%, respectively; difference, 18.7% [95% CI, –13.2% to 50.7%]; P = .31).
Similarly, in the melanoma cohort (n = 1295), more germline pLOF variants validated in the tumor sequencing data were identified by the deep learning method than by the standard method (619 vs 582, respectively), resulting in higher sensitivity (91.7% vs 86.2%; difference, 5.5% [95% CI, 2.2% to 8.8%]; P = .001), lower specificity (30.8% vs 69.2%; difference, –38.4% [95% CI, −43.6% to −33.3%]; P < .001), and lower PPV (59.4% vs 75.6%; difference, –16.2% [95% CI, −20.4% to −11.9%]; P < .001). The NPV for the deep learning method was not significantly different from the standard method (77.0% vs 82.0%, respectively; difference, –5.0% [95% CI, –11.2% to 1.3%], P = .11; Table 2).
Detection of pLOF Variants in 12 Commonly Used Clinical Multigene Panels
Among 286 patients with prostate cancer, the deep learning method vs the standard method identified pLOF variants that were judged to be in the following clinical multigene panels: cardiovascular disorders (36 vs 34, respectively), ciliopathies (43 vs 40), dermatological disorders (24 vs 23), hearing loss (33 vs 33), hematological disorders (38 vs 36), mitochondrial disorders (49 vs 48), neurological disorders (178 vs 173), neuromuscular disorders (33 vs 32), prenatal screening (118 vs 110), pulmonary disorders (19 vs 18), kidney disorders (48 vs 44), and retinal disorders (232 vs 223) (eFigures 9A and 10A in Supplement 1).
In the melanoma cohort (n = 1295), the deep learning method vs the standard method identified pLOF variants that were judged to be valid in the following clinical multigene panels: cardiovascular disorders (45 vs 44, respectively), ciliopathies (34 vs 37), dermatological disorders (19 vs 18), hearing loss (41 vs 39), hematological disorders (31 vs 30), mitochondrial disorders (32 vs 27), neurological disorders (162 vs 155), neuromuscular disorders (30 vs 23), prenatal screening (107 vs 104), pulmonary disorders (17 vs 17), kidney disorders (39 vs 41), and retinal disorders (212 vs 204) (eFigure 9B and 10B in Supplement 1).
Properties of Pathogenic Variants Exclusively Identified by 1 Method
The deep learning method vs the standard method identified 36 vs 19 frameshift variants, respectively, 31 vs 17 stop codon variants, and 40 vs 11 canonical splice-site variants that were judged to be valid (eFigure 11 in Supplement 1). For pathogenic variants exclusively identified using the deep learning method, there were false-positive variants identified in genes with poor sequencing coverage compared with true-positive variants (mean [SD], 7.1 [6.8] reads vs 21.4 [35.6] reads, respectively; P < .001).
In contrast, for pathogenic variants exclusively identified by the standard method, false-positive variants had similarly sufficient sequencing coverage to the true-positive variants (mean [SD], 44.3 [69.6] reads vs 43.1 [59.4] reads, respectively, P = .46; eFigure 12 in Supplement 1). In addition, even though the deep learning and standard methods identified the same number of common variants (minor allele frequency >1%), the mean number of additional rare variants (minor allele frequency <1%) identified by the deep learning method in each patient was 49.6 (95% CI, 46.7 to 52.7) variants per exome in the prostate cancer cohort and 101.2 (95% CI, 95.8 to 106.9) variants per exome in the melanoma cohort (eFigures 13A and 13B in Supplement 1).
Discussion
Analysis of pathogenic variant detection in 2 cohorts of individuals with prostate cancer and melanoma showed that a deep learning method identified more pathogenic variants in cancer-predisposition genes that were judged to be valid (true-positive) than the current standard method, resulting in higher sensitivity, specificity, PPV, and NPV. However, these findings also demonstrated that the deep learning and standard methods were complementary in that the application of both approaches to the sequence data yielded the highest number of pathogenic variants judged to be valid.
Identification of pathogenic variants has substantial clinical implications for pathogenic variant carriers and their at-risk family members. For example, the National Comprehensive Cancer Network recommends offering risk-reducing salpingo-oophorectomy before the age of 50 years to female carriers of pathogenic variants in RAD51D (OMIM: 602954) or BRIP1 (OMIM: 605882),29 similar to those discovered only by the deep learning method used in this analysis. In addition, it is recommended to consider a more intensive breast cancer screening approach (using breast magnetic resonance imaging starting at the age of 40 years) for female carriers of pathogenic germline ATM (OMIM: 607585) variants.29 This clinical actionability also extends to many noncancer pathogenic variants exclusively discovered by the deep learning method, including those in ATP7B (OMIM: 606882) because presymptomatic initiation of chelating therapy can effectively prevent the life-threatening complications of Wilson disease30 and those in the multigene panels because any additional germline analysis yield may translate into more patients benefiting from the clinical utility of establishing a molecular diagnosis.
Overall, these findings suggested that although both methods had comparable performance for detecting common variants, the deep learning method had a higher sensitivity for detecting rare pathogenic variants, an observation that can be explained by examining the underlying approach of each method. The standard method uses joint genotyping, which leverages population-wide information from all analyzed samples and high-quality population-based data sets, such as 1000 Genomes31 and dbSNP,32 to determine the quality of each identified variant. Although this approach enables the standard method to effectively identify variants that are seen frequently in the analyzed and reference genomic data sets (ie, relatively common in the population), joint genotyping, and the subsequent filtering step, called variant quality score recalibration, are inherently biased toward filtering out variants that are very rare (ie, only encountered once in the analyzed data set).
Because 97.3% of the cancer-predisposition variants had an allele frequency of less than 1:10 000,33 exceptionally large patient cohorts are needed to effectively use the power of joint genotyping on these ultrarare variants. Conversely, the deep learning method used in this article evaluates the sequencing images of each variant individually using deep neural networks, thus mimicking the standard workflow of how geneticists assess the evidence supporting genetic variants in each sample independently.13,24 In addition to having a higher sensitivity and specificity compared with the standard method, this sample-based analysis approach also avoids the n + 1 problem for clinical genetics laboratories in which all cohort samples need to be jointly reanalyzed every time a new sample is added to the study. Such an approach has been proven to be extremely impractical, resource intensive, and time-consuming, especially for large research studies such as the gnomAD database.34
Collectively, this analysis of 2367 germline exomes of patients with cancer consistently showed a higher molecular diagnostic yield for deep learning–based germline pathogenic variant analysis compared with standard methods, regardless of the examined gene set. The higher sensitivity of the deep learning–based method may also lead to an improved ability to uncover novel gene-disease associations in already existing genomic data sets. However, the deep learning–based method was not able to detect all manually validated pathogenic variants in the analyzed data sets, thus a hybrid variant detection approach may achieve a higher sensitivity.
Limitations
This study has several limitations. First, this study only included patients with cancer diagnoses, so the performance of the deep learning method may change when used for patients affected by other conditions. Second, this study largely included patients with European ancestry, and further studies are needed to evaluate the molecular diagnostic yield increment in other ancestral groups. Third, this analysis used convenience cohorts with limited available clinical outcomes, so prospective studies are needed to further evaluate the effect of deep learning variant detection on clinical outcomes. Fourth, given the lack of a practical independent validation process for all examined genomic positions, some true pathogenic and pLOF variants could have been potentially missed by both methods.
Fifth, this study used the best practices of the standard method so analysis frameworks using alternative settings, or a modified version of the standard method, may have different performance rates for pathogenic variant detection. Sixth, this study does not evaluate the performance of these methods on genetic data generated using technologies other than the paired-end, short-read Illumina platform. Seventh, although the prostate cancer and melanoma patient cohorts were used to calculate the PPV and NPV, and for whom germline analysis of the cancer-predisposition genes are frequently performed, these patient cohorts are not commonly tested for the noncancer ACMG or OMIM gene sets, so the calculated PPV and NPV for these gene sets may not represent the actual PPV and NPV of the standard and deep learning methods for patients in whom testing these gene sets is indicated. Eighth, the calculated PPV and NPV for these 2 methods highlight the probability of having the molecular genetic change, not the clinical disease, and were calculated using gene sets, so nucleotide-based and gene-based values may differ.
Conclusions
Among a convenience sample of 2 independent cohorts of patients with prostate cancer and melanoma, germline genetic testing using deep learning, compared with the current standard genetic testing method, was associated with higher sensitivity and specificity for detection of pathogenic variants. Further research is needed to understand the relevance of these findings with regard to clinical outcomes.
eMethods
eFigure 1. Technical overview of the preprocessing, variant calling, and variant analysis steps
eFigure 2. Sequencing and quality control (QC) metrics of the prostate cancer (A, B, and C) and melanoma (D, E, and F) cohorts
eFigure 3. Exome-wide germline variant detection in 1072 germline samples of patients with prostate cancer
eFigure 4. Pathogenic cancer-predisposition variants discovered in 1295 patients with melanoma
eFigure 5. A representative pathogenic predisposition variant in the succinate dehydrogenase complex, subunit A (SDHA) gene that was only detected by deep learning but not standard variant calling methodology
eFigure 6. Effect of considering true positive variants as variants that were considered “valid” by all three examiners
eFigure 7. Performance of GATK and DV models to detect pathogenic variants in 151 cancer predisposition and ACMG genes in 1072 patients with prostate cancer
eFigure 8. Performance of GATK and DV models to detect pathogenic variants in 151 cancer predisposition and ACMG genes in 1295 patients with melanoma
eFigure 9. Performance of the deep learning and standard methods to detect pLOF variants that were judged to be valid in 12 clinically oriented multi-gene panels
eFigure 10. Analysis of the molecular diagnostic yield of the current standard method and deep learning, towards detecting putative LOF variants in 12 phenotype-based multi-gene panels
eFigure 11. Characteristics of validated germline pathogenic variants in 151 ACMG and cancer predisposition genes
eFigure 12. Depth of sequencing coverage of pathogenic variants exclusively called by deep learning and the gold-standard method in 151 cancer predisposition and ACMG genes
eFigure 13. Performance of the standard method, GATK, and deep learning, DV, for detecting common, uncommon, and rare variants (minor allele frequency (MAF) of >5%, 1-5%, and <1% respectively)
eTable 3. Definition of the performance metrics and other terms used in this study
eTable 4. Pathogenic and likely pathogenic variants exclusively detected by deep learning in the cancer predisposition genes in 1072 prostate cancer patients
eTable 5. Pathogenic and likely pathogenic variants exclusively detected by the standard method, GATK, in the cancer predisposition genes in 1072 prostate cancer patients
eTable 6. Pathogenic and likely pathogenic variants exclusively detected by the standard method in the cancer predisposition genes in 1295 melanoma patients
eTable 7. Pathogenic and likely pathogenic variants exclusively detected by deep learning in the cancer predisposition genes in 1295 melanoma patients
eTable 8. Pathogenic and likely pathogenic variants exclusively detected by the standard method in the ACMG genes in 1072 prostate cancer patients
eTable 9. Pathogenic and likely pathogenic variants exclusively detected by deep learning in the ACMG genes in 1072 prostate cancer patients
eTable 10. Pathogenic and likely pathogenic variants exclusively detected by the standard method in the ACMG genes in 1295 melanoma patients
eTable 11. Pathogenic and likely pathogenic variants exclusively detected by deep learning in the ACMG genes in 1295 melanoma patients
eTable 12. Performance of the standard and deep learning models in 1072 and 1295 patients with prostate cancer and melanoma using the ACMG and the cancer predisposition gene sets
eNotes
eReferences
eTable 1
eTable 2
References
- 1.Rebbeck TR, Friebel T, Lynch HT, et al. . Bilateral prophylactic mastectomy reduces breast cancer risk in BRCA1 and BRCA2 mutation carriers: the PROSE Study Group. J Clin Oncol. 2004;22(6):1055-1062. doi: 10.1200/JCO.2004.04.188 [DOI] [PubMed] [Google Scholar]
- 2.AlDubayan SH. Leveraging clinical tumor-profiling programs to achieve comprehensive germline-inclusive precision cancer medicine. JCO Precision Oncology. 2019;3:1-3. doi: 10.1200/PO.19.00108 [DOI] [PubMed] [Google Scholar]
- 3.Litton JK, Rugo HS, Ettl J, et al. . Talazoparib in patients with advanced breast cancer and a germline BRCA mutation. N Engl J Med. 2018;379(8):753-763. doi: 10.1056/NEJMoa1802905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Le DT, Durham JN, Smith KN, et al. . Mismatch repair deficiency predicts response of solid tumors to PD-1 blockade. Science. 2017;357(6349):409-413. doi: 10.1126/science.aan6733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.AlDubayan SH, Pyle LC, Gamulin M, et al. ; Regeneron Genetics Center (RGC) Research Team . Association of inherited pathogenic variants in checkpoint kinase 2 (CHEK2) with susceptibility to testicular germ cell tumors. JAMA Oncol. 2019;5(4):514-522. doi: 10.1001/jamaoncol.2018.6477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kemp Z, Turnbull A, Yost S, et al. . Evaluation of cancer-based criteria for use in mainstream BRCA1 and BRCA2 genetic testing in patients with breast cancer. JAMA Netw Open. 2019;2(5):e194428. doi: 10.1001/jamanetworkopen.2019.4428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.AlDubayan SH, Giannakis M, Moore ND, et al. . Inherited DNA-repair defects in colorectal cancer. Am J Hum Genet. 2018;102(3):401-414. doi: 10.1016/j.ajhg.2018.01.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Huang K-L, Mashl RJ, Wu Y, et al. ; Cancer Genome Atlas Research Network . Pathogenic germline variants in 10,389 adult cancers. Cell. 2018;173(2):355-370.e14. doi: 10.1016/j.cell.2018.03.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.DePristo MA, Banks E, Poplin R, et al. . A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet. 2011;43(5):491-498. doi: 10.1038/ng.806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gurovich Y, Hanani Y, Bar O, et al. . Identifying facial phenotypes of genetic disorders using deep learning. Nat Med. 2019;25(1):60-64. doi: 10.1038/s41591-018-0279-0 [DOI] [PubMed] [Google Scholar]
- 11.Esteva A, Robicquet A, Ramsundar B, et al. . A guide to deep learning in healthcare. Nat Med. 2019;25(1):24-29. doi: 10.1038/s41591-018-0316-z [DOI] [PubMed] [Google Scholar]
- 12.Wu Y, Schuster M, Chen Z, et al. Google’s Neural Machine Translation system: bridging the gap between human and machine translation. Published September 26, 2016. Accessed October 13, 2020. https://arxiv.org/abs/1609.08144
- 13.Poplin R, Chang P-C, Alexander D, et al. . A universal SNP and small-indel variant caller using deep neural networks. Nat Biotechnol. 2018;36(10):983-987. doi: 10.1038/nbt.4235 [DOI] [PubMed] [Google Scholar]
- 14.Armenia J, Wankowicz SAM, Liu D, et al. ; PCF/SU2C International Prostate Cancer Dream Team . The long tail of oncogenic drivers in prostate cancer. Nat Genet. 2018;50(5):645-651. doi: 10.1038/s41588-018-0078-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Poplin R, Ruano-Rubio V, DePristo MA, et al. . Scaling accurate genetic variant discovery to tens of thousands of samples. Accessed October 13, 2020. https://www.biorxiv.org/content/10.1101/201178v3
- 16.Bohannan ZS, Mitrofanova A. Calling variants in the clinic: informed variant calling decisions based on biological, clinical, and laboratory variables. Comput Struct Biotechnol J. 2019;17:561-569. doi: 10.1016/j.csbj.2019.04.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lek M, Karczewski KJ, Minikel EV, et al. ; Exome Aggregation Consortium . Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;536(7616):285-291. doi: 10.1038/nature19057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tennessen JA, Bigham AW, O’Connor TD, et al. ; Broad GO; Seattle GO; NHLBI Exome Sequencing Project . Evolution and functional impact of rare coding variation from deep sequencing of human exomes. Science. 2012;337(6090):64-69. doi: 10.1126/science.1219240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zook JM, Chapman B, Wang J, et al. . Integrating human sequence data sets provides a resource of benchmark SNP and indel genotype calls. Nat Biotechnol. 2014;32(3):246-251. doi: 10.1038/nbt.2835 [DOI] [PubMed] [Google Scholar]
- 20.Github website Best practices for multi-sample variant calling with DeepVariant. Accessed July 3, 2020. https://github.com/google/deepvariant
- 21.Geraldine_VdAuwera GATK best practices for variant discovery in DNAseq: GATK forum. Published September 13, 2013. Accessed July 3, 2020. https://gatkforums.broadinstitute.org/gatk/discussion/3238/best-practices-for-variant-discovery-in-dnaseq
- 22.Richards S, Aziz N, Bale S, et al. ; ACMG Laboratory Quality Assurance Committee . Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015;17(5):405-424. doi: 10.1038/gim.2015.30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Barnell EK, Ronning P, Campbell KM, et al. . Standard operating procedure for somatic variant refinement of sequencing data with paired tumor and normal samples. Genet Med. 2019;21(4):972-981. doi: 10.1038/s41436-018-0278-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Robinson JT, Thorvaldsdóttir H, Wenger AM, Zehir A, Mesirov JP. Variant review with the integrative genomics viewer. Cancer Res. 2017;77(21):e31-e34. doi: 10.1158/0008-5472.CAN-17-0337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Loveday C, Turnbull C, Ramsay E, et al. ; Breast Cancer Susceptibility Collaboration (UK) . Germline mutations in RAD51D confer susceptibility to ovarian cancer. Nat Genet. 2011;43(9):879-882. doi: 10.1038/ng.893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ramus SJ, Song H, Dicks E, et al. ; AOCS Study Group; Ovarian Cancer Association Consortium . Germline mutations in the BRIP1, BARD1, PALB2, and NBN genes in women with ovarian cancer. J Natl Cancer Inst. 2015;107(11):djv214. doi: 10.1093/jnci/djv214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Helgason H, Rafnar T, Olafsdottir HS, et al. . Loss-of-function variants in ATM confer risk of gastric cancer. Nat Genet. 2015;47(8):906-910. doi: 10.1038/ng.3342 [DOI] [PubMed] [Google Scholar]
- 28.Bausch B, Schiavi F, Ni Y, et al. ; European-American-Asian Pheochromocytoma-Paraganglioma Registry Study Group . Clinical characterization of the pheochromocytoma and paraganglioma susceptibility genes SDHA, TMEM127, MAX, and SDHAF2 for gene-informed prevention. JAMA Oncol. 2017;3(9):1204-1212. doi: 10.1001/jamaoncol.2017.0223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.National Comprehensive Cancer Network NCCN guidelines for genetic/familial high-risk assessment: breast and ovarian. Accessed October 13, 2020. https://www.nccn.org/about/news/ebulletin/ebulletindetail.aspx?ebulletinid=535
- 30.European Association for Study of Liver EASL clinical practice guidelines: Wilson’s disease. J Hepatol. 2012;56(3):671-685. doi: 10.1016/j.jhep.2011.11.007 [DOI] [PubMed] [Google Scholar]
- 31.1000 Genomes Project Consortium A global reference for human genetic variation. Nature. 2015;526(7571):68-74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sherry ST, Ward M, Sirotkin K. dbSNP—database for single nucleotide polymorphisms and other classes of minor genetic variation. Genome Res. 1999;9:677-679. [PubMed] [Google Scholar]
- 33.Kobayashi Y, Yang S, Nykamp K, Garcia J, Lincoln SE, Topper SE. Pathogenic variant burden in the ExAC database: an empirical approach to evaluating population data for clinical variant interpretation. Genome Med. 2017;9(1):13. doi: 10.1186/s13073-017-0403-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Karczewski KJ, Francioli LC, Tiao G, et al. The mutational constraint spectrum quantified from variation in 141,456 humans. Accessed October 13, 2020. https://www.biorxiv.org/content/10.1101/531210v4 [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
eMethods
eFigure 1. Technical overview of the preprocessing, variant calling, and variant analysis steps
eFigure 2. Sequencing and quality control (QC) metrics of the prostate cancer (A, B, and C) and melanoma (D, E, and F) cohorts
eFigure 3. Exome-wide germline variant detection in 1072 germline samples of patients with prostate cancer
eFigure 4. Pathogenic cancer-predisposition variants discovered in 1295 patients with melanoma
eFigure 5. A representative pathogenic predisposition variant in the succinate dehydrogenase complex, subunit A (SDHA) gene that was only detected by deep learning but not standard variant calling methodology
eFigure 6. Effect of considering true positive variants as variants that were considered “valid” by all three examiners
eFigure 7. Performance of GATK and DV models to detect pathogenic variants in 151 cancer predisposition and ACMG genes in 1072 patients with prostate cancer
eFigure 8. Performance of GATK and DV models to detect pathogenic variants in 151 cancer predisposition and ACMG genes in 1295 patients with melanoma
eFigure 9. Performance of the deep learning and standard methods to detect pLOF variants that were judged to be valid in 12 clinically oriented multi-gene panels
eFigure 10. Analysis of the molecular diagnostic yield of the current standard method and deep learning, towards detecting putative LOF variants in 12 phenotype-based multi-gene panels
eFigure 11. Characteristics of validated germline pathogenic variants in 151 ACMG and cancer predisposition genes
eFigure 12. Depth of sequencing coverage of pathogenic variants exclusively called by deep learning and the gold-standard method in 151 cancer predisposition and ACMG genes
eFigure 13. Performance of the standard method, GATK, and deep learning, DV, for detecting common, uncommon, and rare variants (minor allele frequency (MAF) of >5%, 1-5%, and <1% respectively)
eTable 3. Definition of the performance metrics and other terms used in this study
eTable 4. Pathogenic and likely pathogenic variants exclusively detected by deep learning in the cancer predisposition genes in 1072 prostate cancer patients
eTable 5. Pathogenic and likely pathogenic variants exclusively detected by the standard method, GATK, in the cancer predisposition genes in 1072 prostate cancer patients
eTable 6. Pathogenic and likely pathogenic variants exclusively detected by the standard method in the cancer predisposition genes in 1295 melanoma patients
eTable 7. Pathogenic and likely pathogenic variants exclusively detected by deep learning in the cancer predisposition genes in 1295 melanoma patients
eTable 8. Pathogenic and likely pathogenic variants exclusively detected by the standard method in the ACMG genes in 1072 prostate cancer patients
eTable 9. Pathogenic and likely pathogenic variants exclusively detected by deep learning in the ACMG genes in 1072 prostate cancer patients
eTable 10. Pathogenic and likely pathogenic variants exclusively detected by the standard method in the ACMG genes in 1295 melanoma patients
eTable 11. Pathogenic and likely pathogenic variants exclusively detected by deep learning in the ACMG genes in 1295 melanoma patients
eTable 12. Performance of the standard and deep learning models in 1072 and 1295 patients with prostate cancer and melanoma using the ACMG and the cancer predisposition gene sets
eNotes
eReferences
eTable 1
eTable 2


