Figure 2. Examples of Germline Pathogenic Variants That Were Exclusively Identified by the Deep Learning or Standard Variant Detection Methods.
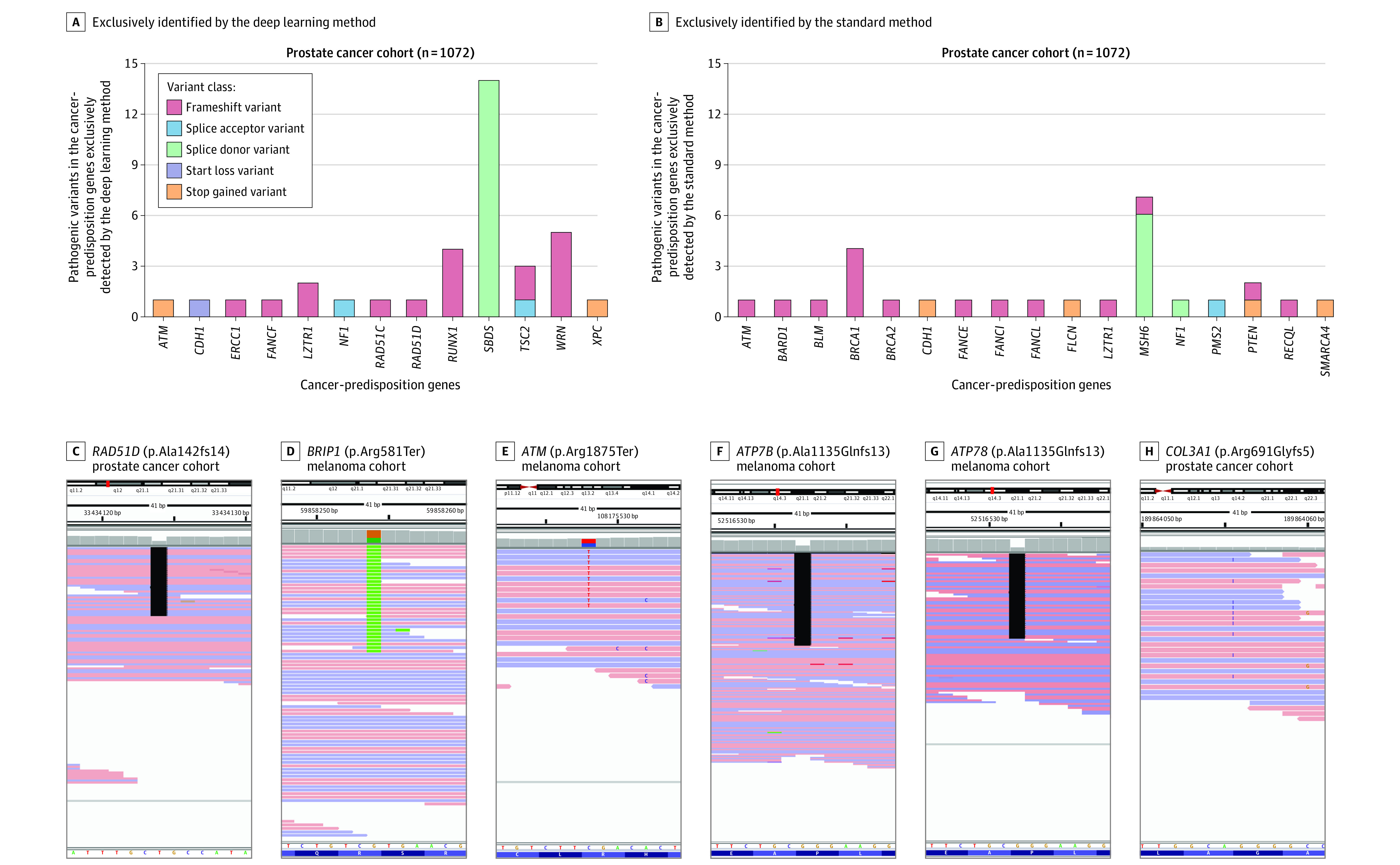
Thirty-six germline pathogenic variants were identified by the deep learning method, 75% of which were judged to be valid or true-positive (A), whereas 27 variants were identified by the standard method, 40.7% of which were judged to be valid (B). A similar analysis of 1295 patients with melanoma showed a similarly higher detection rate with the deep learning method (eFigure 4). Examples show pathogenic variants in the cancer-predisposition gene set (C-E) and the clinically actionable American College of Medical Genetics and Genomics gene set (F-H) that were only identified by the deep learning method and judged to be valid on manual assessment.
