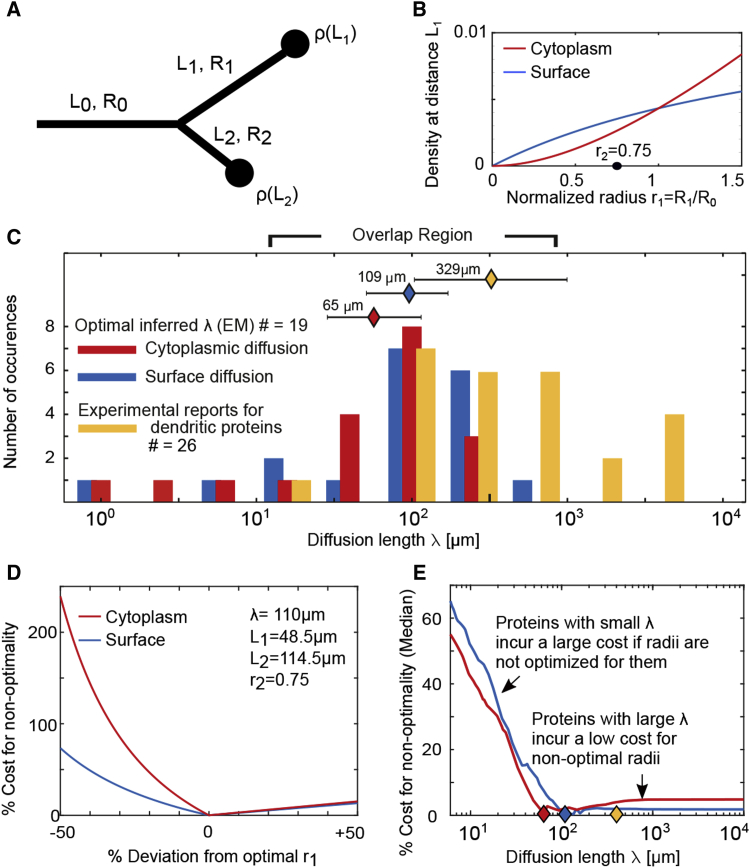
Figure 5.
Daughter Radii in Pyramidal Neurons Are Optimized for Protein Diffusion Lengths that Overlap with the Experimentally Reported Distribution of Dendritic Proteins
(A) A sketch of a dendritic branch and its corresponding radii, lengths, and density of proteins at the tips of the daughter dendrites.
(B) Predicted protein density at the tips of the daughter dendrites as a function of the daughter radius . The second daughter radius is set to the median radius for PPC pyramidal neurons , , and diffusion length .
(C) Distribution showing the number of branches optimized for a particular diffusion length for cytoplasmic (red) and surface (blue) proteins. Shown in yellow are diffusion lengths for 26 dendritic proteins with experimentally reported diffusion coefficients and half-lives (STAR Methods). The median values of the distributions are , , and , respectively.
(D) For a given set of λ, , and , there is only one value of the normalized radius (here, ) that minimizes the total amount of proteins needed to ensure at least proteins are localized to the tips of both daughter branches. A normalized radius of less than the optimal leads to a substantial increase in the number of proteins (cost) needed to populate the dendritic arbor, whereas a higher value does not significantly increase the cost.
(E) Proteins with small diffusion lengths (short-diffusivity proteins) incur a larger cost for non-optimal daughter radii than those with large diffusion lengths.
Experimental methods underlying the data can be found in the STAR Methods sections Dendrite Diameter Measurement in Three-Dimensional Electron Microscopy, Hippocampal Neuron Preparation, Neuron Transfection, and Image Acquisition.