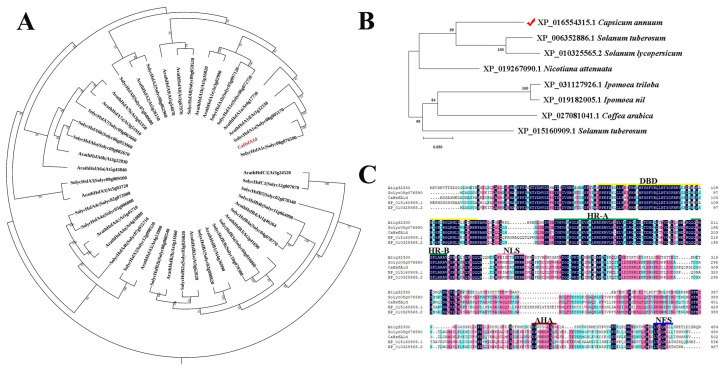
Figure 1.
Sequence analysis of the CaHsfA1d protein. (A) Phylogenetic relationship of the CaHsfA1d with Arabidopsis (ArathHsfs) and tomato (SolycHsfs) Hsf proteins. The Hsf proteins of the Arabidopsis and tomato were downloaded from the Heatster (http://www.cibiv.at/services/hsf/). The neighbor-joining (NJ) phylogenetic tree was constructed using MEGA-X (http://www.megasoftware.net/) with the following parameters: Poisson model, bootstrap (1000 replicates), and pairwise deletion gaps. (B) Phylogenetic relationship of CaHsfA1d (XP_016554315.1) and Hsfs from other plant species. The protein sequence of CaHsfA1d was used to perform the BLASTP search in the database NCBI Protein Reference Sequences (https://blast.ncbi.nlm.nih.gov/). Eight HSF proteins were selected to construct the NJ phylogenetic tree by MEGA-X using the parameters in (A). (C) Multiple sequence alignment of Hsf proteins. Clustal Omega software (http://www.clustal.org/) was used to align the Hsf sequences using default parameters and the results were minor repaired by DNAMAN Version 9.0 (www.lynnon.com) software. The domains have been indicated. At1g32330 (Arabidopsis thaliana, ArathHsfA1d), Solyc08g076590 (Solanum lycopersicum, SolycHsfA1c), XP_015160909.1 (Solanum tuberosum), XP_010325565.2 (Solanum lycopersicum).