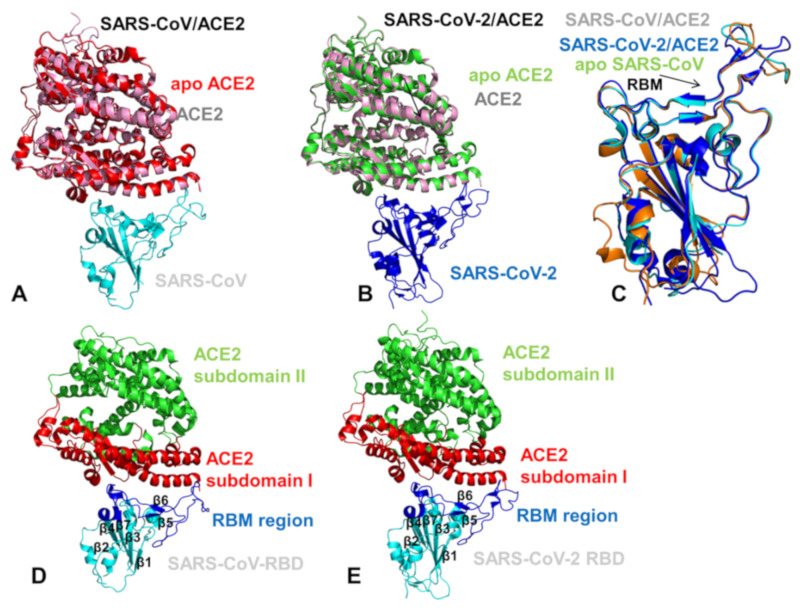
Figure 1.
Structural organization of the SARS-CoV-RBD (severe acute respiratory syndrome coronavirus receptor binding domain) and SARS-CoV-2-RBD (severe acute respiratory syndrome coronavirus 2 receptor binding domain) complexes with human ACE enzyme. (A) A general overview of the SARS-CoV-RBD complex with ACE2 (pdb id 2AJF). The SARS-CoV-RBD is in cyan ribbons and the bound ACE2 enzyme is in pink ribbons. The crystal structure of the unbound native human ACE2 enzyme (pdb id 1R42) is superimposed and shown in red ribbons. (B) A general overview of the SARS-CoV-2-RBD complex with ACE2 (pdb id 6M0J). The SARS-CoV-2-RBD is shown in blue ribbons, the bound ACE2 enzyme is in pink ribbons, and the unbound form of ACE2 is in green. (C) Structural superposition of the unbound SARS-CoV-RBD (pdb id 2GHV) (in orange), bound SARS-CoV-RBD (pdb id 2AJF) (in cyan), and SARS-CoV-2-RBD (pdb id 6M0J) (in blue). Structural similarity of the RBM motif is indicated by an arrow and annotated. A moderate local mobility of the flexible ridge loop at the tip of the RBM in the unbound and bound SARS-CoV-RBD forms is evident. (D) A general overview of the secondary structure elements and binding interface in the SARS-CoV-RBD complex with human ACE2. The SARS-CoV-RBD is shown in cyan and secondary structure elements are annotated. The RBM region in SARS-CoV-RBD (residues 424–494) that provides the contact interface with ACE2 is highlighted in blue ribbons and annotated. The subdomain I of human ACE2 is shown in red ribbons and the subdomain II is shown in green ribbons. (E) A general overview of the secondary structure elements and binding interface in the SARS-CoV-2 RBD complex with human ACE2. The SARS-CoV-RBD is shown in cyan and secondary structure elements are annotated. The RBM region is in blue ribbons and annotated. The subdomains I and II of human ACE2 are shown in red and green ribbons, respectively.