Figure EV1. The principle steps of our algorithm Revelio for extracting the cell cycle from the data.
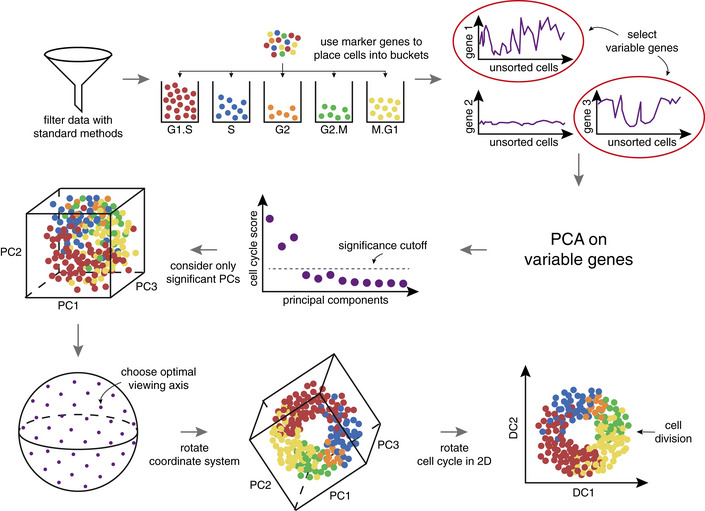
After the data are filtered by standard methods, we divide the cells into buckets with the help of marker genes (Whitfield et al., 2002; Macosko et al., 2015). Next, we select variable genes (Butler et al., 2018) and apply PCA on the reduced data set. Afterward, we utilize a cell cycle marker and cluster score (Materials and Methods) to judge which PCs are influenced by the cell cycle. The significant PCs are used to construct three‐dimensional subspaces. We then choose an optimal viewing axis by minimizing the cell cycle score along the viewing axis (Materials and Methods). The coordinate system is rotated linearly and the cell cycle is obtained only within the plane spanned by the first two new axes (DC1, DC2). Within the DC1‐DC2 plane, we estimate the time point of cell division and rotate the cell cycle plane accordingly (Materials and Methods).
