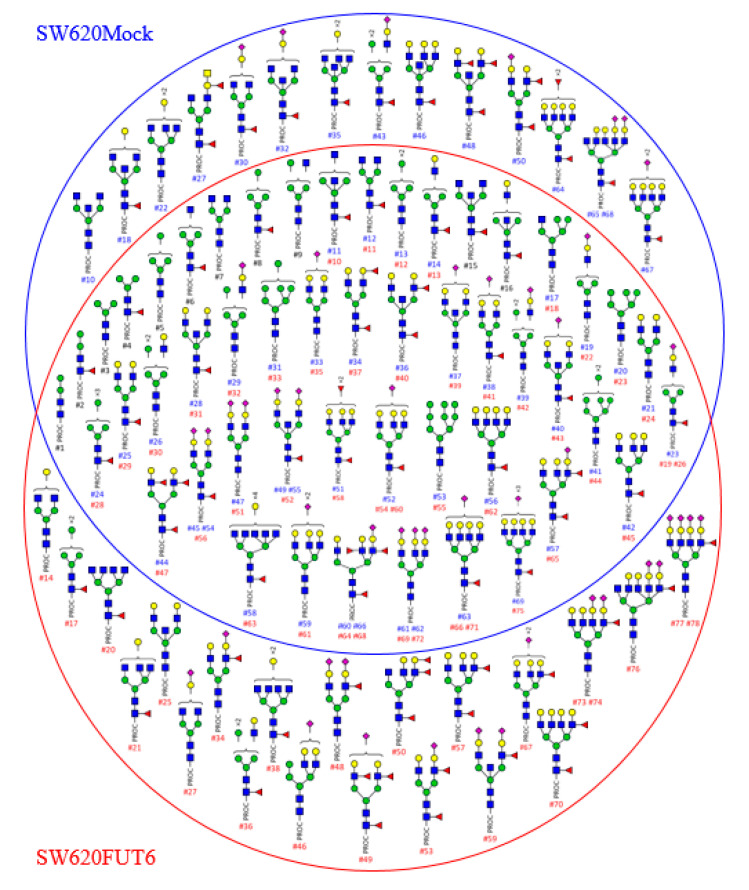
Figure 2.
N-glycans identified by HILIC-UHPLC-MS from membrane proteins of SW620Mock and FUT6 cells. Schematic representation of the N-glycans released from SW620Mock cells (inside the blue circle) and from SW620FUT6 cells (inside the red circle) membrane proteins. Structures identified in both cell lines are shown where the two circles overlap. Sixty-nine N-glycan structures were identified for SW620Mock membrane proteins and 78 for SW620FUT6 membrane proteins. Y- and B-ion fragments identification, which allowed N-glycan composition determination, is depicted in Supplementary Materials Tables S1 and S2 for SW620Mock and SW620FUT6 cells membrane proteins, respectively. In the Supplementary Materials Tables S1 and S2, a peak ID has been attributed to each structure, which corresponds to the #number (#) under each N-glycan (blue for N-glycans from SW620Mock proteins, red for N-glycans from SW620FUT6 proteins and black for identical peak ID number attributed to N-glycans from both SW620Mock and FUT6 proteins). Structures are depicted with the following notation: PROC: procainamide; blue square: N-acetylglucosamine; green circle: Mannose; yellow circle: Galactose; red triangle: Fucose; purple diamond: N-acetylneuraminic acid.