Table 3.
Summary of branch model comparisons
Branch models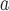
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|
| Scenarios of adaptive evolution | Single episode preceding C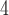 evolution evolution |
Single episode during C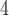 evolution, at the base of Andropogoneae evolution, at the base of Andropogoneae |
Two episodes during C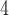 evolution, at the base of Arundinellinae and Andropogoneae s.s. evolution, at the base of Arundinellinae and Andropogoneae s.s.
|
||||||
Gene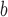
|
N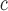
|
Null model | Basal branch | Sustained | Basal branch | Sustained | Basal branches | Sustained | dN/dS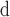
|
Core C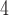 genes genes |
|||||||||
| nadpmdh-1P1 (NADP-MDH) | 28 | 13.53 | 14.82 | 5.90 | 14.74 | 0.00* | 14.55 | 1.46* | 0.07–0.14 |
| nadpme-1P4 (NADP-ME) | 63 | 205.29 | 202.70 | 42.20* | 205.26 | 0.00* | 202.73 | 2.89* | 0.08–0.30 |
| pck-1P1 (PCK) | 41 | 36.77 | 38.11 | 5.40* | — | — | 38.77 | 0.00* | 0.02–0.06 |
| ppc-1P3 (PEPC) | 51 | 105.82 | 107.82 | 7.83* | 82.81* | 0.00* | 85.87* | 21.78* | 0.03–0.09 |
| ppdk-1P2 (PPDK) | 30 | 100.33 | 97.23 | 51.07* | 83.78* | 37.69* | 0.00* | 53.91* | 0.10–0.44 |
Paralogs of C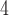 genes genes |
|||||||||
| nadpme-1P1 | 30 | 1.52 | 1.56 | 0.00 | 1.28 | 1.16 | 3.33 | 1.73 | 0.09 |
| nadpme-1P2 | 21 | 0.00 | 0.72 | 1.59 | 2.00 | 0.97 | 0.55 | 0.93 | 0.09 |
| nadpme-1P3 | 24 | 10.58 | 0.00* | 4.73 | 4.77 | 7.75 | 9.97 | 10.05 | 0.05–0.17 |
| ppc-1P4 | 30 | 3.92 | 4.08 | 5.90 | 5.23 | 5.30 | 0.00 | 3.84 | 0.07 |
| ppc-1P5 | 30 | 0.00 | 2.00 | 1.84 | 1.83 | 1.49 | 1.84 | 1.80 | 0.06 |
| ppc-1P7 | 19 | 11.33 | 13.29 | 5.59 | — | — | 12.65 | 0.00* | 0.09–0.04 |
| ppdk-1P1 | 12 | 6.19 | 8.17 | 6.31 | 7.39 | 0.37 | 0.00 | 0.63 | 0.18 |
| Other nuclear genes | |||||||||
| arodeh | 30 | 0.00 | 1.99 | 1.70 | — | — | 1.85 | 1.77 | 0.11 |
| dwarf8 | 39 | 46.51 | 45.75 | 23.99* | 47.23 | 2.35* | 48.34 | 0.00* | 0.06–0.16 |
| knotted1 | 13 | 4.15 | 0.00 | 5.45 | — | — | 3.60 | 5.94 | 0.07 |
| phyB | 55 | 7.95 | 4.67 | 8.35 | 0.00 | 9.86 | 9.71 | 9.85 | 0.09 |
| waxy | 55 | 0.00 | 1.74 | 1.70 | — | — | 1.90 | 1.91 | 0.05 |
Note: The best-fit model is highlighted in bold. Asterisks indicate significant likelihood ratio tests (LRT) against the null model after Bonferroni correction. Two hypotheses of potential enzyme adaptation were tested for each scenario, the first assuming a shift in selective pressure only in the basal branch(es) of the group specified (‘Basal branch’), the second assuming a sustained shift from the basal branch including all descendant branches (‘Sustained’). Missing values correspond to trees in which Andropogoneae was not monophyletic.
 Delta-AIC values relative to the best-fit model for each gene are shown.
Delta-AIC values relative to the best-fit model for each gene are shown.
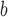 C
C gene annotation following Moreno-Villena et al. (2018).
gene annotation following Moreno-Villena et al. (2018).
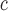 Number of sequences in the alignment.
Number of sequences in the alignment.
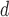 dN/dS ratios of background and foreground branches, respectively, estimated for the best-fit model, except in cases where the null was the best-fit model, for which there was a single dN/dS estimate for all branches.
dN/dS ratios of background and foreground branches, respectively, estimated for the best-fit model, except in cases where the null was the best-fit model, for which there was a single dN/dS estimate for all branches.
