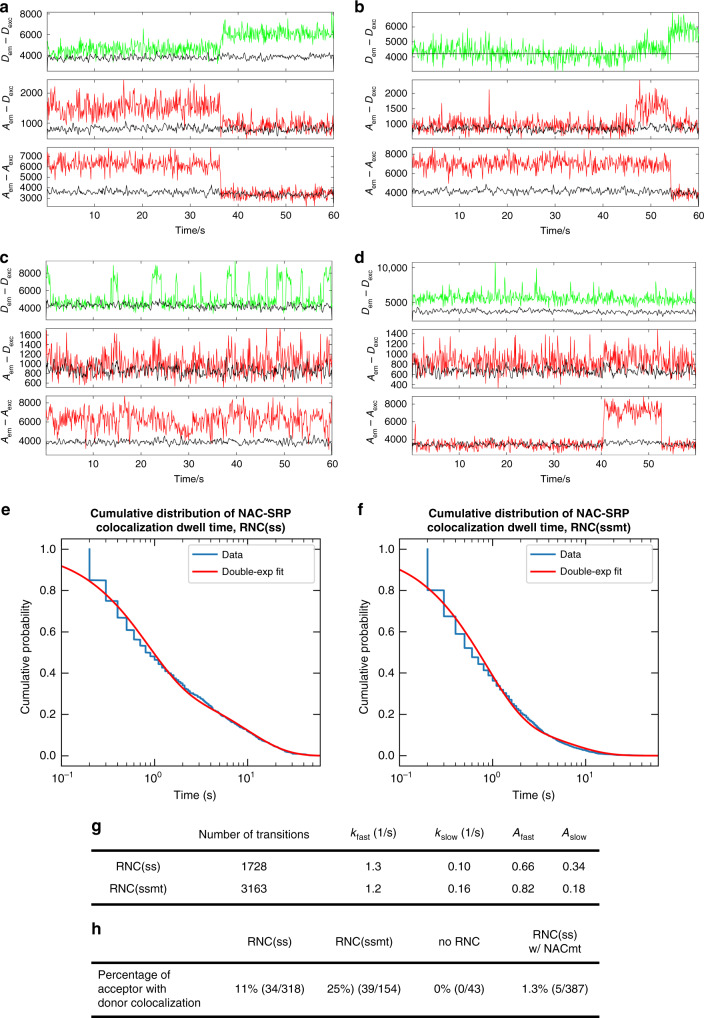
Fig. 5. Single-molecule colocalization of NAC and SRP on surface immobilized RNCs.
a–d Representative traces of NAC-SRP colocalization on surface immobilized RNC(ss) (a, b) and RNC(ssmt) (c, d). Biotinylated quartz surface was coated with 1.5 nM purified monosome RNC(ss) or RNC(ssmt) with 3’ biotinylated mRNA. The sample chamber was then flushed with 2 nM NACCy3B and 1 nM SRPAtto647n (labeled at SRP54-S12C) in image buffer. Movies were recorded in ALEX mode for 60 s at a speed of 10 frames per second. Note the anti-correlation in Aem–Dex and Dem–Dex panels that indicates FRET between NACCy3B and SRPAtto647n. e–g Dwell time analysis of the NAC binding events to RNC(ss) (e) and RNC(ssmt) (f). Single molecule fluorescence traces were fit to a Hidden Markov Model (HMM) to extract dwell times of NAC in the colocalized state. The cumulative distributions of dwell times were fitted to a double-exponential function Eq. (17) in the Method, and the fitted parameters are reported in (g). Number of transition is the total number of transitions observed for NAC binding to and dissociation from the RNC-SRP complex. h Summary of the frequency of observed SRP-NAC colocalization events under each condition, calculated from the ratio of the number of acceptors with colocalized donor over the total number of acceptors detected. No RNC indicates that 2 nM NACCy3B and 1 nM SRPAtto647n (labeled at 54-S12C) were incubated in image buffer on microscope slides without immobilized RNC. RNC(ss) w/ NACmt is the same as RNC(ss) except that 2 nM of NACmt instead of NAC was used.