Figure 2.
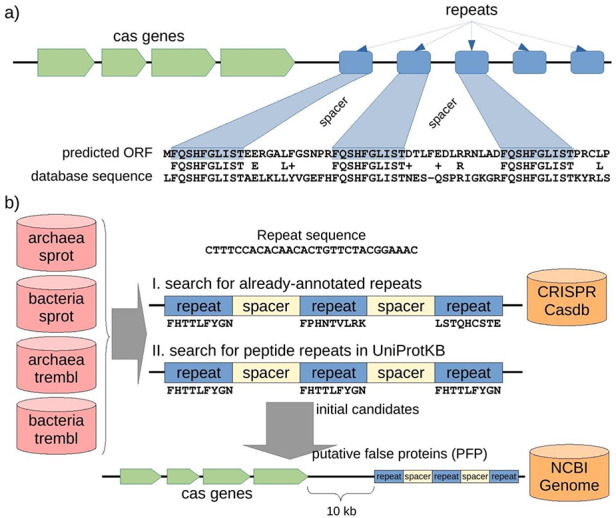
Structure of a CRISPR-Cas locus and its appearance when it is translated, and protocol to discover misannotated CRISPR sequences in protein databases. (a) A CRISPR-Cas locus includes a series of cas protein-coding genes followed by short nucleotide repeats surrounding heterogeneous sequences of a similar length called spacers. When a CRISPR sequence is erroneously translated, the corresponding amino acid sequence could present repeats separated by uniform spacers, and this protein would show similarity of 50% (centered in the repeat region) with other spurious proteins. (b) Searching for putative spurious sequences originating from translated CRISPR in four subsets of the UniProtKB protein database. The first approach (I) consisted in searching for translations of repeat sequences from the CRISPRCasdb database separated by putative spacers. The second approach (II) consisted in searching for amino acid repeats separated by putative spacers directly in the protein sequences. Finally, the initial candidates from the two approaches were mapped to their corresponding genomic sequences, and cas genes were searched within 10 kb around the candidate (see Methods for details). Proteins with both, repeats and cas genes nearby, are expected to be originating from the translation of spurious ORFs from CRISPR sequences, so called putative false proteins (PFP). Note that the first approach can take into account three different peptide sequences (drawn from the three possible reading frames of the nucleotide repeat sequence), while the second approach can only take into account one peptide sequence for all the possible repeats but can potentially discover new CRISPR repeats other than those already-annotated in the CRISPR database.
