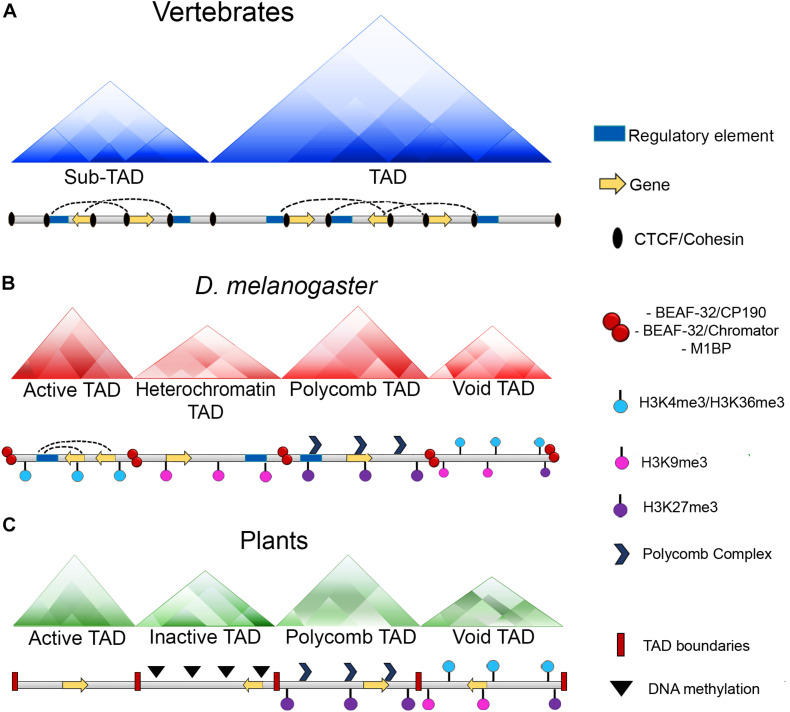
FIGURE 2.
Chromatin organization among different organisms. (A) In vertebrates, chromatin loops are formed and stabilized by the presence of CTCF and the cohesin complex. Moreover, TAD boundaries present an enrichment of CTCF and the cohesin complex which act as insulators and keep these domains detached. (B) In Drosophila, chromatin loops exist, however, there is not an enrichment of the dCTCF ortholog with the cohesin complex at the boundaries of these domains, instead, this role it is carried out by other architectural proteins of the fly as BEAF-32, CP190, chromator or M1BP. Additionally, in Drosophila TADs can be classified according to their epigenetic states: Active TADs which possess an enrichment of active histone marks as H3K4me3 and H3K36me3; heterochromatin TADs which have an enrichment of repressive marks as H3K9me3; Polycomb TADs which are enriched with the presence of Polycomb complexes and the H3K27me3 mark and void TADs which do not have a defined landscape. (C) In plants, CTCF is not conserved and there is not a report of any protein with insulator activities. However, the existence of TAD-like domains has been reported and as well as Drosophila, these domains can be classified in four distinct categories which are: Active TADs; inactive TADs characterized by a high degree of DNA methylation; Polycomb TADs and void TADs.