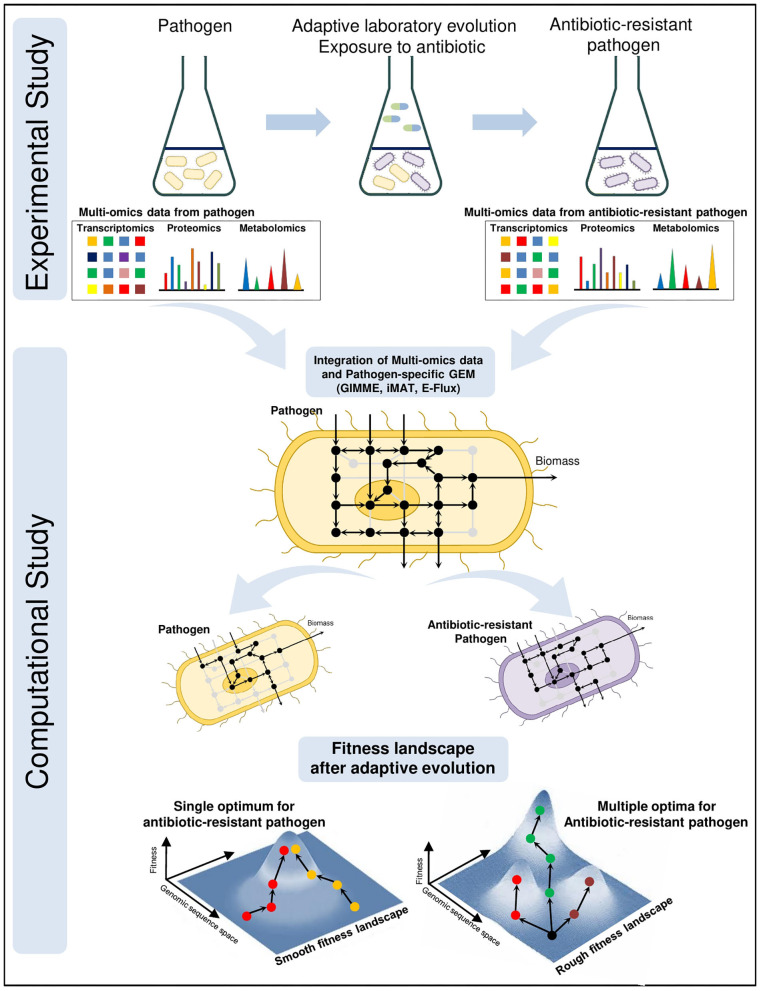
Figure 2.
Integrated investigation of the antibiotic resistance by adaptive laboratory evolution (ALE) experiments and pathogen-specific GEMs. Multi-omics data are collected during the ALE experiments from wild-type and antibiotic-resistant pathogens at different time points. To elucidate the evolutionary response at system-level due to antibiotic pressure, high throughput omics data are computationally mapped onto the genome-scale metabolic network. Analysis of metabolic shift in the cellular metabolism and mechanism of antibiotic-resistant pathogenic GEMs facilitate the discovery of novel potential drug targets and treatment strategies against antibiotic-resistant pathogen. Fitness landscapes demonstrate the optimality in adaptive evolution of antibiotic resistance. Smooth fitness landscapes consist of a single optimum and regardless of the starting point evolutionary tendency converge to this optimum. There exist multiple optima on the rough fitness landscapes and evolutionary tendency diverge even from the same starting point.