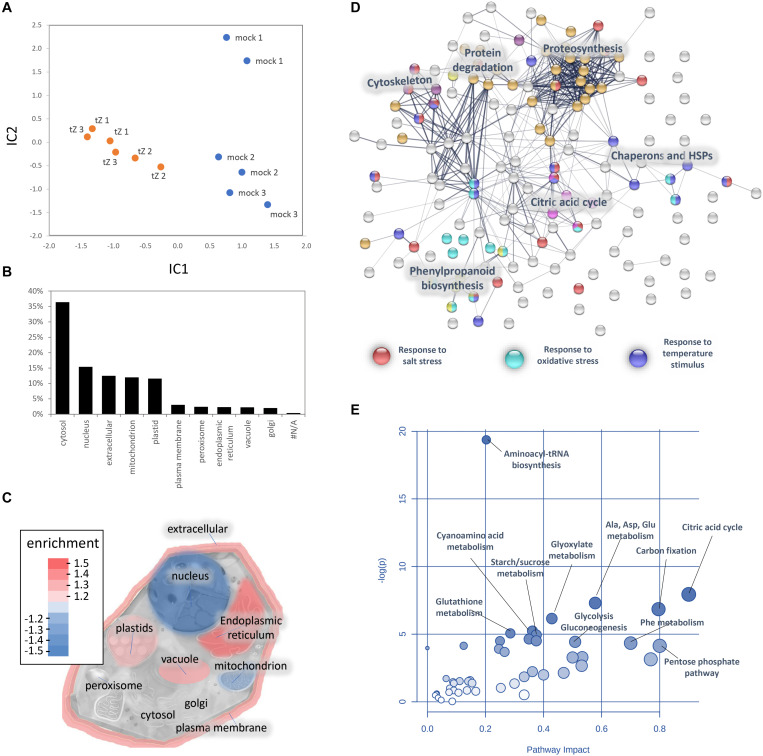
FIGURE 3.
Cytokinin impact on the barley root proteome and metabolome. (A) The proteome profile separation of cytokinin- and mock-treated samples. Independent component analysis based on quantitative data of 972 proteins. Results of three biological replicates represent proteins with at least 10 matched spectra in a biological replicate and more than > 95% of the detected root proteome. (B) Expected localization of quantified barley root proteins and (C) visualization of organellar enrichment in cytokinin-responsive proteins. Localization based on data prediction by CropPal and SUBA4.0 (Hooper et al., 2016, 2017); (D) Interactions and functional clusters of cytokinin-responsive proteins highlighted by String (Szklarczyk et al., 2019). The selected highlighted categories are represented by at least five and ten proteins for ‘Biological Processes’ and ‘KEGG Pathways,’ respectively; (E) Cytokinin impact on barley root metabolism. Highlighted pathways were identified by an integrative analysis of identified cytokinin-responsive proteins and metabolites (MetaboAnalyst; Pang et al., 2020). Results (D-E) are based on data available for putative Arabidopsis orthologs.