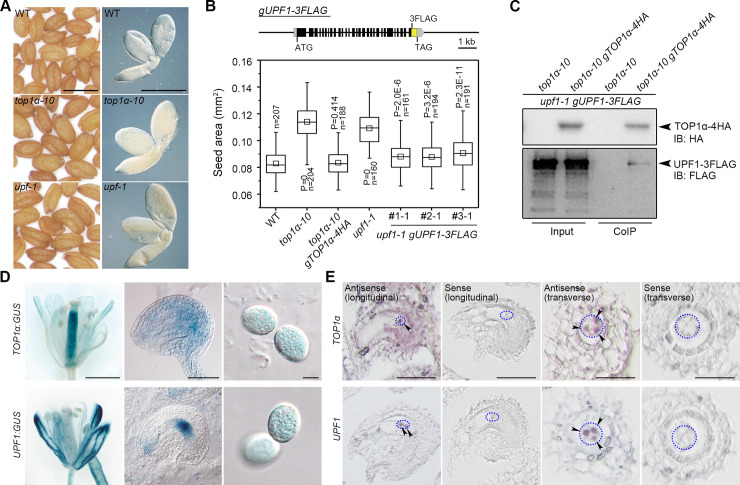
Fig 1. TOP1α and UPF1 regulate seed size and interact in the nucleus.
(A) top1α-10 and upf1-1 produce larger seeds than WT. Left panels, mature seeds; right panels, mature embryos. Scale bars, 0.5 mm. (B) Seed sizes of top1α-10 and upf1-1 are rescued by correspondent genomic fragments. Upper panel: schematic diagram of the gUPF1-3FLAG construct. Lower panel: seed sizes of different genetic backgrounds. P values were determined by two-tailed Mann-Whitney U-test compared to WT. (C) In vivo interaction between TOP1α and UPF1 shown by CoIP. Nuclear protein extracts from pistils of the specified genotypes were immunoprecipitated by anti-HA antibody. The input and CoIP proteins were detected by anti-HA (upper panel) and anti-FLAG antibody (lower panel), respectively. (D) Representative GUS staining of TOP1α:GUS and UPF1:GUS in opening flowers (left panels), ovules (middle panels), and pollen grains (right panels). Scale bars, 1 mm, 50 μm, 10 μm from left to right. (E) In situ localization of TOP1α and UPF1 mRNA expression in ovules. Blue dotted ellipses indicate the regions where antipodal cells are located, and arrowheads indicate antipodal cells with signals. Scale bars, 50 μm, 50 μm, 25 μm, and 25 μm from left to right. The data underlying this figure are included in S1 Data and S1 Raw Images. CoIP, co-immunoprecipitation; GUS, β-glucuronidase; HA, hemagglutinin; n, number of seeds examined; IB, immunoblotting; WT, wild type.