Abstract
There is increasing awareness of the role genetic risk variants have in mediating vulnerability to psychiatric disorders such as schizophrenia and autism. Many of these risk variants encode synaptic proteins, influencing biological pathways of the postsynaptic density and, ultimately, synaptic plasticity. Fragile-X mental retardation 1 (FMR1) and cytoplasmic fragile-X mental retardation protein (FMRP)-interacting protein 1 (CYFIP1) contain 2 such examples of highly penetrant risk variants and encode synaptic proteins with shared functional significance. In this review, we discuss the biological actions of FMRP and CYFIP1, including their regulation of (i) protein synthesis and specifically FMRP targets, (ii) dendritic and spine morphology, and (iii) forms of synaptic plasticity such as long-term depression. We draw upon a range of preclinical studies that have used genetic dosage models of FMR1 and CYFIP1 to determine their biological function. In parallel, we discuss how clinical studies of fragile X syndrome or 15q11.2 deletion patients have informed our understanding of FMRP and CYFIP1, and highlight the latest psychiatric genomic findings that continue to implicate FMRP and CYFIP1. Lastly, we assess the current limitations in our understanding of FMRP and CYFIP1 biology and how they must be addressed before mechanism-led therapeutic strategies can be developed for psychiatric disorders.
Keywords: Fragile-X mental retardation protein, Cytoplasmic FMRP-interacting protein 1, Fragile-X mental retardation protein targets, Synapse, Psychiatric disorders
The Synapse and Postsynaptic Density Proteins
Neurotransmission between presynaptic and postsynaptic terminals is the adaptive communication mechanism linking neurons and other cell types into neural circuits and networks, which form the basis of synaptic plasticity, cognition, and behaviour [1, 2]. The majority of excitatory, glutamatergic synapses in the mammalian brain are located at small dendritic protrusions, or spines [3], and contain a prominent assembly of proteins at the postsynaptic membrane known as the postsynaptic density (PSD) [4, 5]. Proteomic profiling of the PSD has revealed over 1,000 different proteins [6, 7, 8], many of which converge on the regulation of synaptic plasticity through biological pathways controlling protein synthesis, receptor trafficking, or structural rearrangements [9, 10, 11].
Synaptic FMRP: Regulator of mRNA and Local Translation
One such synaptic protein is fragile-X mental retardation protein (FMRP), encoded by the FMR1 gene (Xq27.3) [12] and the monogenic cause of neurodevelopmental disorder fragile X syndrome (FXS) [13]. FMR1 mRNA is expressed in the neuronal cell body, developing and mature axons, dendrites and dendritic spines, and the nucleus [14, 15], but not across all neuronal populations [16, 17]. FMRP is an RNA-binding protein with multiple structural motifs for binding RNA (such as the K homology domain and arginine-glycine-rich, RGG, box) [18], capable of regulating the dendritic sequestering, and localization, of hundreds of target neuronal mRNAs [19, 20], either through direct interactions or via intermediary interactions with noncoding RNA [21, 22].
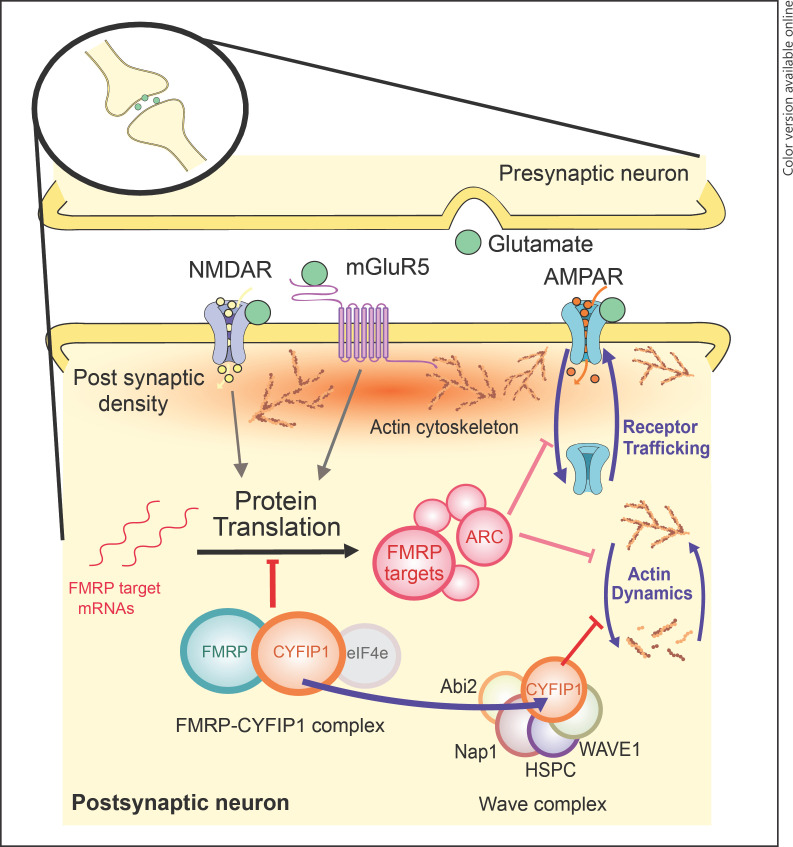
FMRP, its target mRNA, and other protein partners together form large messenger ribonucleoparticles (mRNPs) [23]. Within the mRNPs, FMRP plays a key role in the translational silencing of its target mRNA [24, 25, 26, 27], required during the transport of mRNA along dendrites [28], before synaptic activation results in the docking of the mRNPs to the spines and subsequent translation [29, 30]. FMRP specifically regulates the rate-limiting step of cap-dependent mRNA translation initiation by binding to the initiation factor eIF4E and FMRP-binding partner cytoplasmic FMRP-interacting protein 1 (CYFIP1) (see section “Synaptic CYFIP1: A Negative Regulator of Protein Synthesis and Cytoskeletal Dynamics” later) [31, 32], although initiation may also be regulated via FMRP ubiquitination or sumoylation [33, 34, 35]. FMRP also controls elongation stages of mRNA translation by stalling ribosomes on FMRP target transcripts [19, 24], although how FMRP switches between the regulation of initiation and elongation is currently unknown [36]. FMRP is, therefore, a critical mediator of local translation of mRNA targets, acting at both presynaptic and postsynaptic terminals [24, 37, 38]. Some of the key biological roles of FMRP at postsynaptic terminals are highlighted in Figure 1.
Fig. 1.
Biological roles of synaptic FMRP and CYFIP1 in postsynaptic neurons. FMRP plays a key role in negatively regulating the translation of hundreds of FMRP targets, including activity-regulated cytoskeleton associated (ARC), by forming a complex with CYFIP1, alongside the initiation factor eIF4e. The control of mRNA translation, and its repression by the CYFIP1-FMRP complex, is partly mediated through activation of upstream NMDA and mGluR5 receptors. FMRP targets such as ARC can drive changes in synaptic plasticity through regulation of α-Amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid (AMPA) receptor trafficking/internalization and increasing actin cytoskeleton stability. Meanwhile, CYFIP1 can bind and inhibit the WAVE regulatory complex, thereby blocking the promotion of actin cytoskeleton rearrangements. Preclinical evidence suggests that under conditions of synaptic activation, CYFIP1 redistributes between the 2 main complexes, with greater association with the WAVE complex and a reciprocal decrease with the FMRP complex. FMRP, fragile-X mental retardation protein; CYFIP1, cytoplasmic FMRP-interacting protein; mGluR5, metabotropic glutamate receptor 5.
Beyond translational silencing, FMRP plays other biological roles [39], including RNA editing [40], regulation of mRNA target stability [41], and ion-channel binding [42, 43], collectively influencing calcium signalling [44], activity-dependent neurodevelopment [45], and the balance of excitatory/inhibitory circuits [46, 47]. The additional functions of FMRP might explain instances where FMRP does not appear to be a straightforward repressor of protein synthesis [48], perhaps most pertinently through FMRP's ability to influence the stability of a subset of mRNAs [22, 49].
FMRP Targets
Considerable effort has been made to identify the mRNAs targeted by FMRP so that biological pathways affected by mutations in FMR1 can be better predicted. Such studies have used immunoprecipitation followed by either microarray [50, 51] or high-throughput sequencing [19, 52] to determine FMRP-bound mRNAs. Due to varied methodology and tissues (as well as influence from type I and II errors), these lists of FMRP targets differ considerably [52, 53]. Therefore, precisely which mRNAs are bound by FMRP is uncertain and likely to be somewhat tissue-specific. Two studies using mouse cortical tissue and comparable methodology (high-throughput sequencing of RNA isolated by cross-linking immunoprecipitation) yielded highly overlapping results: 89% of mRNAs identified as FMRP targets by Darnell and colleagues [19] were also identified by Maurin et al. [52]. Still, only a small subset of the proposed targets have been validated [23, 49, 54, 55, 56, 57].
Gene ontology analyses of brain-derived FMRP targets confirm an overrepresentation of genes involved in functions related to synaptic activity, plasticity, development, and anatomy [19, 52, 58], consistent with studies of FMRP function. The proteins they encode include both presynaptic and postsynaptic components. Of these are subunits and interactors of receptor complexes considered central to synaptic plasticity phenotypes associated with FXS, chiefly the of metabotropic glutamate receptor 5 (mGluR5) and N-methyl-D-aspartate (NMDA) receptor signalling complexes [19]. The observation that FMRP binds some presynaptic proteins supports the evidence that FMRP regulates protein synthesis during axon development and synapse formation [59, 60, 61, 62].
Whilst studies of FMRP targets have identified probable interactions between FMRP and ribosomal mRNAs, further work is needed to determine whether the translation of these mRNAs is indeed repressed by FMRP within the regulatory complex together with CYFIP1 and eIF4E proteins in the cell [31, 32].
Synaptic CYFIP1: A Negative Regulator of Protein Synthesis and Cytoskeletal Dynamics
CYFIP1 is a highly dynamic synaptic protein involved in numerous biological pathways through an array of protein-protein interactions (Fig. 1) [63]. Originally known as specifically Rac1-activated protein 1 (SRA-1) [64], CYFIP1 was later found to bind with FMRP [12, 65], forming a critical CYFIP1-FMRP complex at the synapse [31]. Specifically, FMRP-bound CYFIP1 acts as a non-canonical eIF4E-binding protein [31], thereby providing competition for the binding of eIF4E to the translation initiation complex (eIF4E-eIF4G) [66, 67]. Overall, it is this eIF4E-CYFIP1-FMRP complex, together with its target mRNA, that represses translation at dendritic and synaptic sites [31]. Upon synaptic activation via tropomyosin receptor kinase B or group I mGluRs, eIF4E is released from CYFIP1-FMRP and permits the translation of target mRNAs [31]. A subsequent study has implicated a mitogen-activated protein (MAP)-kinase-interacting kinase-dependent pathway in the release of the inhibitory CYFIP1-FMRP complex from target mRNA, via MAP-kinase-interacting kinase phosphorylation of CYFIP1, in the early phase of long-term potentiation (LTP), thereby permitting translation [68].
Aside from its role in regulating protein synthesis, CYFIP1 forms part of the ∼400-kDa heteropentameric WAVE regulatory complex, which also contains WAVE1/2/3, abl interactor-1/2 (ABI1/2), Nck-associated protein 1 (NCKAP1), and haematopoietic stem/progenitor cell protein 300 (HPSC300) components [69]. Without CYFIP1, the WAVE complex promotes actin cytoskeleton remodelling via the Arp2/3 complex [70, 71, 72], impacting on aspects of dendritic spine formation, stability, morphology, migration, and excitability [73]. The role of CYFIP1 is to maintain the WAVE complex in an inhibited state, until the GTPase, Rac1, causes the dissociation of CYFIP1 from the WAVE regulatory complex and allows actin remodelling to proceed via Arp2/3 [74].
CYFIP1 belongs to the two aforementioned complexes, FMRP and WAVE, in a mutually exclusive manner, skewed towards greater association with the WAVE complex, under basal conditions [74]. Notably, synaptic activation changes the protein conformation of CYFIP1, from globular to planar, and drives the distribution of CYFIP1 further towards the RAC1-WAVE complex, with a concomitant decrease in the eIF4E-CYFIP1 complex [71, 74, 75]. Therefore, CYFIP1 is a central molecular mediator that bridges the 2 processes of mRNA translation and actin dynamics, both essential for synaptic plasticity [76, 77, 78]. Other molecular roles for CYFIP1 are being explored, including its role presynaptically. For instance, presynaptic function is altered in the hippocampus of juvenile Cyfip1 knockout (KO) mice, thought to derive from changes in presynaptic terminal size and enhanced vesicle release probability, and driven by dysregulation of the WAVE complex [79]. These findings closely align with previous findings in cyfip1 mutant fly models that specifically found alterations in actin polymerization in presynaptic terminals [65, 80]. More recently, Cyfip1 KO mice were found to have decreased myelination of callosal axons, alongside impaired presynaptic neurotransmission in the corpus callosum [81].
CYFIP1 has a closely related paralogue, CYFIP2, with over 88% amino acid identity [12]. Like CYFIP1, CYFIP2 is found at excitatory [82] and inhibitory [83] synapses, and binds both to the WAVE complex [63, 69] and to FMRP [12]. Interestingly, CYFIP2 additionally binds to FMRP-related proteins, FXR1P and FXR2P, while CYFIP2 mRNA is a target of FMRP [19], implying a further layer of feedback between FMRP and the family of CYFIPs. However, the molecular redundancy between these paralogues is limited, given that the deletion of both copies of CYFIP1 is embryonically lethal [74, 84]. Furthermore, CYFIP1, but not CYFIP2, has been consistently associated with neuropsychiatric disorders [82, 83; however, see 85].
Psychiatric Disorders and the Synapse
Considerable evidence suggests that a wide range of neuropsychiatric disorders, such as FXS, autism spectrum disorders (ASDs), schizophrenia, intellectual disabilities (IDs), and bipolar disorder, exhibit convergent synaptic pathology [6, 32, 86, 87, 88, 89, 90, 91]. Synaptic dysfunction has been observed at several levels, including genetic alterations [92, 93, 94],aberrant proteins [95] and their translation [25, 96, 97, 98], molecular signalling pathways [99, 100, 101], spine morphology [102], aberrant synaptic plasticity [103, 104, 105], neurocircuitry, and connectivity [106]. These interrelated observations highlight impaired synaptic function as a common feature of several neuropsychiatric disorders [91, 94, 107].
In light of this view, and the biological importance of FMRP and CYFIP1 at the synapse (outlined in sections “Synaptic FMRP: Regulator of mRNA and Local Translation” and “Synaptic CYFIP1: A Negative Regulator of Protein Synthesis and Cytoskeletal Dynamics”), we will now consider the role of FMRP and CYFIP1 in the aetiology of psychiatric disorders, using data from human patient studies, especially psychiatric genomics, and preclinical models.
FMRP and FMRP Targets in Psychiatric Disorders
FXS Patients and Fmr1 KO Models
In humans, transcriptional silencing of the FMR1 gene by a triplet repeat expansion (beyond 200 repeats, typically ∼800) in the 5-untranslated region of FMR1[108] leads to FXS [13, 109]. FXS patients display a broad range of abnormalities, including increased immaturity of dendritic spines [110, 111, 112], altered molecular signalling [23], increased levels of basal protein synthesis [113, 114], altered neuron and circuit excitability [115], structural and connectivity defects in brain networks [116], and a range of cognitive and behavioural phenotypes that overlap considerably with ID and ASD [117, 118, 119]. Indeed, FXS represents the single most common form of inherited ID with a prevalence of 1:4,000 males and 1:8,000 females [120] and the most common, single-gene cause of ASD [108, 117]. FMRP may also be involved in other neuropsychiatric disorders, beyond FXS and related ASDs, including schizophrenia and bipolar disorder [121, 122, 123, 124, 125].
The effects of Fmr1 mutations have been interrogated preclinically for 25 years through the Fmr1 KO mouse model [126] and, with the advent of modern gene-targeting technologies, the Fmr1 KO rat model [127, 128]. Many of the features of human FXS have been recapitulated in Fmr1 KO mouse and rat models, especially in 3 key areas: dendritic spine maturation [112, 127, 129, 130], elevated basal protein synthesis [127, 131, 132, 133], and behavioural/cognitive phenotypes, including ASD-like abnormalities [134], abnormalities in social interaction and interest [135], social anxiety [136], and reduced behavioural flexibility/reversal learning in a variety of tasks [127, 137, 138, 139, 140, 141].
In addition to heightened global protein synthesis, Fmr1 KO rodents display a lack of mGluR-dependent translational control, which results in an elevated protein synthesis-dependent form of synaptic plasticity, known as mGluR-mediated long-term depression [127, 142, 143, 144, 145]. Increased mGluR-dependent translation is thought to occur through excessive activation of the mGluR5 subtype, given that reductions in mGluR5 expression [132], or the mGluR5 antagonist 2-Methyl-6-(phenylethynyl)pyridine (MPEP)[46, 146], can rescue several Fmr1 KO phenotypes. The altered mGluR5 signalling in the absence of Fmr1 appears to be mediated through the preferential interaction of mGluR5 with activity-dependent isoforms of Homer1 over constitutive Homer proteins [147, 148].
The deletion of Fmr1 results in the loss of the repressive eIF4E-Cyfip1-FMRP complex, which de-represses the initiation complex, eIF4F, required for cap-dependent translation initiation of FMRP targets [149]. It was shown that an inhibitor of the eIF4F complex, which creates free eIF4E, increases the abundance of the eIF4E-CYFIP1-FMRP complex (with a parallel decrease in the CYFIP1-WAVE complex) in Fmr1 KO mice, and the restoration of this imbalance rescues spine and memory deficits in these animals [150]. Hence, studies of the Fmr1 KO rodent model have illuminated a variety of molecular mechanisms relevant to FXS, especially those pertinent to the regulation of protein synthesis, and may provide biological targets for therapeutic intervention [24], complementing ongoing clinical trials in human FXS patients [151, 152].
FMRP and FMRP Targets in Psychiatric Genomic Studies
Beyond repeat expansions in the FMR1 gene, a number of rare pathogenic point mutations have been reported that cause developmental delay and ID reminiscent of FXS [153, 154, 155, 156, 157]. Further evidence suggests that mutations in the autosomal homolog FXR2 gene might also contribute to ID [158, 159, 160]. Whilst variants affecting the related FXR1 gene confer risk to schizophrenia, bipolar disorder, and autism [161, 162, 163, 164, 165], the genetic link between FMR1 and psychiatric disorders derives from enrichment of association within the gene targets of FMRP (among which the fragile-X family genes themselves are included).
A set of FMRP target mRNAs derived from a study of mouse cortical polyribosomes [19] have been recurrently highlighted in the literature due to their enrichment for genes associated with an array of psychiatric disorders. Through large-scale genome-wide association studies, these 842 FMRP targets have been shown to be genetically associated with schizophrenia [161, 162], autism [166], major depressive disorder [167], and bipolar disorder [58]. In addition to the risk conferred from common variation, this gene set is enriched for rare variants from patients with schizophrenia [168, 169, 170, 171], autism [172], and bipolar disorder [173]; de novo variants from patients with schizophrenia [174] and autism [175, 176, 177]; and to a lesser extent copy number variants from patients with schizophrenia [178, 179, 180]. The convergence of risk from multiple different types of genetic variants forms a strong evidence base, implicating this gene set in psychiatric pathology. Conversely, FMRP targets derived from a study of human embryonic kidney cells [51] do not appear to be associated with psychiatric disorders [166, 168], highlighting the tissue specificity of these relationships.
Brain FMRP targets overlap considerably with other gene sets associated with psychiatric disorders, such as genes encoding PSD proteins and those involved in calcium signalling, synaptic plasticity, learning, and memory [19, 52, 58, 181]. However, despite these overlaps, the enrichment of brain FMRP targets for association with psychiatric disorders is independent [58, 162, 168] and proportional to the confidence of binding by FMRP [58]. Moreover, in many instances, it appears that FMRP targets capture subsets of these other gene sets in which genetic association is concentrated [58]. Hence, this set of genes locally regulated by FMRP during plasticity and development at the synapse may represent a collection of biological pathways important for the manifestation of a range of psychiatric disorders.
CYFIP1 in Psychiatric Disorders
15q11.2 Copy Number Variants and Cyfip1 Dosage Models
The proximal long arm of human chromosome 15 (15q11.2–13.3) is a region of numerous low-copy repeats that can lead to aberrant meiotic chromosomal rearrangements. These result in deletions or duplications of sections of DNA, known as copy number variants (CNVs), and occur at any of 5 common breakpoints (BP1–BP5) on chromosome 15 [182]. Neurodevelopmental psychiatric disorders Prader-Willi syndrome and Angelman syndrome are caused by deletions of paternal or maternal origin, respectively, and occur as either large deletions (type I, between BP1 and BP3) or smaller deletions (type II, between BP2 and BP3). Meanwhile, CYFIP1 is cytogenetically positioned in the non-imprinted 500-kb region between BP1 and BP2 on chromosome 15 (15q11.2 interval), along with 3 additional genes: non-imprinted in Prader-Willi/Angelman 1 (NIPA1) and 2 (NIPA2), and tubulin gamma complex-associated protein (TUBGCP5) [183]. The 15q11.2 chromosomal region was first implicated with neurodevelopmental psychiatric disorders through the observation that type I deleted Prader-Willi syndrome or Angelman syndrome patients, who lack the 15q11.2 interval, had more severe behavioural phenotypes than type II deleted patients, in whom the 15q11.2 interval is intact [184, 185]. Later, patients were identified with deletions and duplications between BP1 and BP2, which specifically flanked the 15q11.2 interval itself [182].
Deletions or duplications of 15q11.2 are present in 1 of 100 people who present for genetic screening, whilst incidence in the general population is likely to be nearer 1 in 500 people [186]. The CNV causes patients to display language/motor deficits or delays, behavioural problems, autism, and seizures [187, 188, 189], with deletions being the most impactful on cognition [187] and referred to as Burnside-Butler syndrome [182, 186]. It was recently observed that 15q11.2 deletion patients have structural and functional changes in the brain that likely relate to the accompanying cognitive phenotypes, including a smaller left fusiform gyrus and altered activation in the left fusiform and the left angular gyri using functional magnetic resonance imaging [190]. In subsequent diffusion tensor imaging studies, 15q11.2 deletion carriers show increased fractional anisotropy [191], indicating alterations in the white matter microstructure [192]. In keeping with these findings, white matter changes in 15q11.2 deletion patients closely mirror the phenotypes of FXS patients [193], suggesting a common pathogenic pathway derived from disruption of CYFIP1-FMRP complexes. Although the 15q11.2 deletion is not fully penetrant, as a significant proportion of the general population are healthy carriers with no overt phenotypes [194], it is likely that subclinical cognitive phenotypes exist even in these “healthy” carriers [195].
Among the genes located within the 15q11.2 locus, CYFIP1 is widely regarded as the most likely to confer the biological and behavioural phenotypes associated with 15q11.2 BP1–BP2 CNVs [84, 191]. This is due, in part, to its known functional association with the FXS-relevant protein FMRP (see sections “Synaptic FMRP: Regulator of mRNA and Local Translation,” “FMRP Targets,” “Synaptic CYFIP1: A Negative Regulator of Protein Synthesis and Cytoskeletal Dynamics,” and “FMRP and FMRP Targets in Psychiatric Disorders”) [31, 74]. Furthermore, the expression of CYFIP1 and components of the WAVE complex is disrupted in patients carrying 15q11.2 deletions [196]; iPSCs derived from these patients exhibit cellular phenotypes mediated by the CYFIP1-WAVE complex [197]; and the knockdown of CYFIP1, specifically, in human progenitor cells alters cytoskeletal remodelling [198]. However, the biological roles of the three remaining genes within the 15q11.2 interval requires further delineation, as, like CYFIP1, they are all expressed in the central nervous system and their expression is altered in patients with 15q11.2 CNVs [199].
Great strides have been made in understanding the consequences of altered Cyfip1 dosage through a variety of in vitro and in vivo rodent preclinical models. For instance, the heterozygous deletion of Cyfip1 in mice results in changes in dendritic and spine morphology [74, 82], which are similarly observed in a forebrain-specific conditional homozygous KO model [83], whilst the overexpression of Cyfip1 also impinges on dendrite and spine morphology [82, 200]. Meanwhile, Cyfip1 appears to affect protein synthesis under basal and activity-dependent conditions. The knockdown of Cyfip1 in cortical neurons in vitro increases the translation of FMRP target, activity-regulated cytoskeleton associated (ARC), under basal conditions and also ablates the activity-dependent translation of ARC, using brain-derived neurotrophic factor treatment to mimic synaptic activation [74]. Similar findings were reported in vivo using Cyfip1 heterozygous KO mice, whereby brain-derived neurotrophic factor treatment was insufficient to release the Cyfip1-FMRP complex from eIF4E, preventing the formation of the eIF4F complex, which subsequently prevented activity-dependent translation of ARC protein [68].
Measures of synaptic plasticity in preclinical models of altered Cyfip1 dosage have revealed elevated levels of mGluR-mediated long-term depression, which become disassociated from mRNA translation pathways [84] − findings that are reminiscent of Fmr1 KO rodent models [127, 144]. Overexpressing Cyfip1 in CA1 hippocampal neurons can lead to increased excitatory neurotransmission, and a concomitant decrease in gamma aminobutyric acid (GABA)ergic neurotransmission at inhibitory synapses, shifting the overall excitation/inhibition balance towards excessive excitation [83]. The same study also showed that the conditional, homozygous KO of Cyfip1 in CA1 hippocampal neurons increased inhibitory GABAergic neurotransmission, along with increased expression of GABA receptors, suggesting a shift of excitation/inhibition balance towards greater inhibition [83]. However, in the haploinsufficient Cyfip1 mouse model, which better models the reduced dosage of CYFIP1 in 15q11.2 deletion patients, GABAergic signalling remains unaltered in the hippocampal dentate gyrus [201].
Brain connectivity and white matter architecture appear to be especially sensitive to reduced Cyfip1 dosage. In Cyfip1 heterozygous KO mice, bilateral connectivity was shown to be reduced across multiple brain regions using resting-state functional magnetic resonance imaging [81]. These alterations were likely due to changes in corpus callosal white matter architecture, measured by (i) a decrease in fractional anisotropy using diffusion tensor imaging and (ii) altered levels of myelination and presynaptic function. Furthermore, many of the white matter phenotypes, including decreased fractional anisotropy, were mirrored in a comparable rat model of Cyfip1 haploinsufficiency [202]. However, it is currently unclear why fractional anisotropy might be decreased in rodent models of reduced Cyfip1 dosage but increased in 15q11.2 deletion patients. This will require further study and may alter our current perception of the effect of CNVs of the 15q11.2 interval.
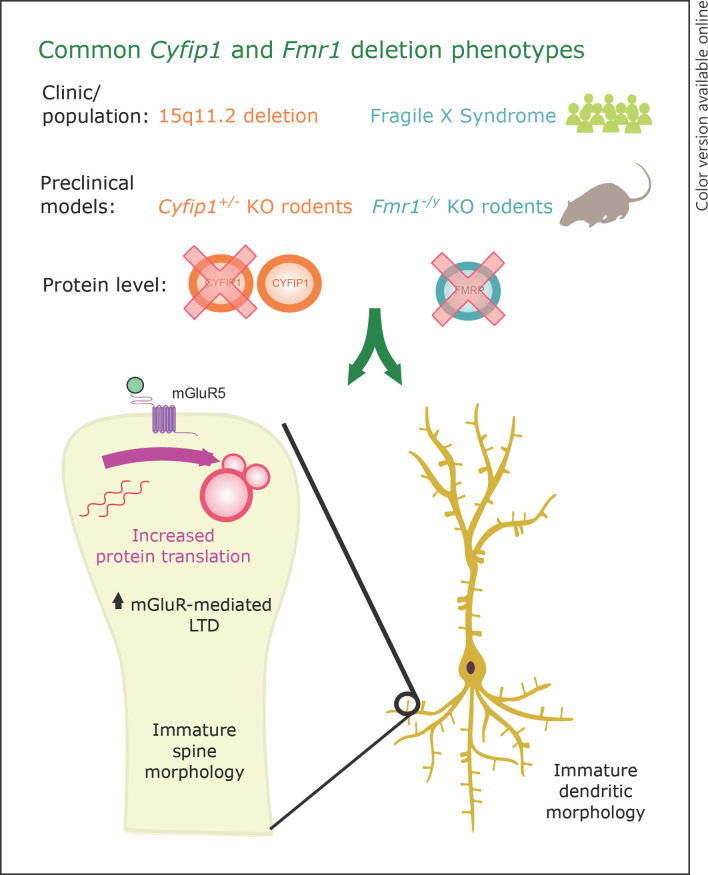
In vivo models of altered Cyfip1 dosage also offer the chance to thoroughly assess changes in behaviour and cognition, prominent features in 15q11.2 deletion (and duplication) patients. Bozdagi and colleagues [84] were the first to behaviourally assess Cyfip1 haploinsufficient mice, and found many aspects of spatial and fear learning and memory to be intact, with the exception of a rapid loss of extinction memory assessed using the inhibitory avoidance paradigm. Subsequent analysis of Cyfip1 heterozygous KO mice and rats has shown specific deficits in motor learning [81, 203], sensorimotor gating measured by prepulse inhibition [81], and behavioural flexibility [202]. Meanwhile, the overexpression of Cyfip1 results in cellular phenotypes, particularly at the dendritic level [200], but appears to have little effect on behaviour and cognition, with the exception of exaggerated fear responses [204]. Overall, there is accumulating evidence that altering the dosage of Cyfip1 in preclinical models leads to profound alterations in cellular and plasticity phenotypes, alongside mild behavioural phenotypes, many of which not only overlap with FXS and the Fmr1 KO model (Fig. 2), but also closely match the key clinical phenotypes of patients with chromosomal deletions (and duplications) of the CYFIP1-containing 15q11.2 interval.
Fig. 2.
Core set of overlapping phenotypes from preclinical Fmr1 and Cyfip1 deletion models. Rodent models of Fmr1 deletion (Fmr1−/y, whereby the single X-linked copy of Fmr1 is deleted in males) or heterozygous Cyfip1 deletion (Cyfip1+/−) mirror clinical populations with FXS and 15q11.2 CNV deletions, respectively. Moreover, these 2 rodent models share a core set of functionally related neurobiological phenotypes, including (i) altered spine and dendritic morphology, (ii) dysregulated protein translation and (iii) elevated long-term depression. Further work is required to fully delineate the consequences of Fmr1 and Cyfip1 deletion, and characterize the similarities. FXS, fragile X syndrome; CNV, copy number variant.
CYFIP1 Variants in Psychiatric Genomic Studies
Genomics studies in psychiatric populations have implicated the 15q11.2 BP1–BP2 deletion with a wide range of psychiatric, neurodevelopmental disorders, including a 2- to 4-fold increased risk for schizophrenia [205, 206], a finding that has been replicated in many subsequent studies [92, 179, 207, 208, 209, 210]. Additionally, 15q11.2 deletions, and duplications, predispose individuals to a 5-fold risk of epilepsy [211], developmental and ID [212, 213, 214], attention deficit hyperactivity disorder [215], major depression [216], and autism [187, 217] [for further review, see 182, 186]. Meanwhile, common variants in CYFIP1 have been reported to increase the risk for ASD [218, 219]. Consistent with the genetic findings, proteomic analysis of prefrontal cortex post-mortem tissue from schizophrenia patients revealed altered levels of CYFIP1 and other proteins belonging to protein synthesis pathways [220].
The relevance of CYFIP1 to schizophrenia becomes especially apparent when considered in the wider context of its biological actions within protein complexes. CYFIP1 is involved in the regulation of ARC protein and ARC-related genes, sometimes referred to as the “ARC complex” (a gene ontology-based complex). CYFIP1 was first associated with schizophrenia in studies that showed an enrichment of the ARC complex (containing 25 genes, of which CYFIP1 is one) in de novo CNV deletions from patients with schizophrenia [92]. The genetic association of this ARC complex with schizophrenia has subsequently been confirmed by exome sequencing studies that assessed single nucleotide variants (SNVs) and indels [168, 174] and larger studies of CNV deletions [178, 179]. Furthermore, the genetic association with schizophrenia of FMRP targets (section “FMRP and FMRP Targets in Psychiatric Genomic Studies”), which are regulated by the CYFIP1-FMRP complex, lends additional evidence to the relevance of CYFIP1 to schizophrenia.
Summary of Findings and Future Directions
FMRP and CYFIP1 are hubs for several biological pathways critical to synaptic plasticity. From preclinical models, we know that reduced expression of either CYFIP1 or FMRP results in a set of core phenotypes: altered spine and dendritic morphology, dysregulated protein synthesis, and elevated long-term depression. A further layer of complexity is added when it is considered that the concerted action of FMRP and CYFIP1, as part of the CYFIP-FMRP complex, represses the translation of hundreds of FMRP targets, likely influencing multiple downstream pathways. The importance of this system to synaptic function is recurrently highlighted by genetic studies demonstrating the risk conferred to psychiatric disorders by variants affecting genes encoding CYFIP1, FMRP, and their targets.
Nevertheless, there are many questions that still surround the biology of FMRP, CYFIP1, and FMRP targets in health and disease. For example, whilst FMRP synaptic biology is well-characterized and preclinical techniques can reverse disorder-relevant phenotypes [132, 151], attempts to move these therapies into the clinic have been largely ineffective [152]. This suggests that further mechanistic insights into the actions of FMRP are needed, alongside further refinement of therapeutic targets and/or strategies. Similarly, whilst FMRP targets are a disease-relevant group of mRNAs, their precise identity and biological function remain underexplored. Meanwhile, the study of CYFIP1 has seen unprecedented advances in recent years, revealing an extensive array of synaptic roles, far beyond its initial characterization as a binding partner to FMRP. Despite the rapid expansion of CYFIP1 studies, many fundamental questions remain and can be addressed in future studies, aided by advances in RNA sequencing, genetic-editing, and proteomic technologies. Whilst extensively characterized, it is also worth noting that the behavioural phenotypes in models of Fmr1 and Cyfip1 deletion are only broadly similar, and in some cases diametrically opposed [221]. These behavioural discrepancies could reflect the diversity of biological function, but might also derive from highly transient and localized interactions between these two proteins.
Penetrant risk variants affecting this biological pathway increase psychiatric vulnerability to a range of psychiatric disorders. For example, CNVs affecting CYFIP1 predispose carriers to increased risk for schizophrenia (mainly 15q11.2 deletions), autism, and ID (mainly 15q11.2 duplications), and likewise FMR1 deletions predispose carriers to autism and ID. These apparently pleiotropic effects might suggest that the categorical nature of diagnoses for psychiatric disorders needs to be fundamentally re-evaluated. Indeed, at the clinic, there are many common patient symptoms that span across diagnostic categories, and patients often present with comorbidities. The genomic findings point towards a continuum of causality, whereby common biological mechanisms, influenced by a range of convergent genetic factors, span across the traditional diagnostic boundaries of psychiatric disorders. The highly tractable mechanism of CYFIP1-FMRP and the regulation of ARC are one such biological pathway, offering a unique entry point for continued study and phenotypic rescue. Future development of novel mechanism-based therapeutic approaches will be vital to meet the ever-growing need to treat these common, yet debilitating, psychiatric disorders.
Disclosure Statement
The authors have no conflicts of interest to declare.
Funding Sources
The work was supported by a Wellcome Trust Strategic Award (DEFINE, 100202/Z/12/Z) and a Medical Research Council Centre Grant (GO801418). S.T. and N.E.C. were supported by Cardiff University Neuroscience and Mental Health Research Institute Fellowships. S.T. has been additionally supported by startup funds associated with a new appointment as lecturer at Keele University.
Author Contributions
The review was conceived by S.T. and written by S.T. and N.E.C.; K.L.T., L.S.W., and J.H. provided additional text, comments, and support. Figures were created by S.T. and N.E.C.
Acknowledgements
The authors would like to acknowledge core support received from the Neuroscience and Mental Health Research Institute, Cardiff University, UK, and School of Life Sciences, Keele University, UK, whilst writing this review.
References
- 1.Ho VM, Lee JA, Martin KC. The cell biology of synaptic plasticity. Science. 2011;334((6056)):623–8. doi: 10.1126/science.1209236. Available from: http://www.ncbi.nlm.nih.gov/pubmed/22053042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Südhof TC. Towards an understanding of synapse formation. Neuron. 2018 Oct;100((2)):276–93. doi: 10.1016/j.neuron.2018.09.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Penzes P, Cahill ME, Jones KA, VanLeeuwen JE, Woolfrey KM. Dendritic spine pathology in neuropsychiatric disorders. Nat Neurosci. 2011;14((3)):285–93. doi: 10.1038/nn.2741. Available from: http://www.ncbi.nlm.nih.gov/pubmed/21346746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhu J, Shang Y, Zhang M. Mechanistic basis of MAGUK-organized complexes in synaptic development and signalling. Nat Rev Neurosci. 2016 Apr;17((4)):209–23. doi: 10.1038/nrn.2016.18. [DOI] [PubMed] [Google Scholar]
- 5.Dosemeci A, Weinberg RJ, Reese TS, Tao-Cheng JH. The postsynaptic density: there is more than meets the eye. Front Synaptic Neurosci. 2016;8:23. doi: 10.3389/fnsyn.2016.00023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bayés A, van de Lagemaat LN, Collins MO, Croning MD, Whittle IR, Choudhary JS, et al. Characterization of the proteome, diseases and evolution of the human postsynaptic density. Nat Neurosci. 2011;14((1)):19–21. doi: 10.1038/nn.2719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sheng M, Hoogenraad CC. The postsynaptic architecture of excitatory synapses: a more quantitative view. Annu Rev Biochem. 2007;76:823–47. doi: 10.1146/annurev.biochem.76.060805.160029. [DOI] [PubMed] [Google Scholar]
- 8.Bayés A, Collins MO, Croning MD, van de Lagemaat LN, Choudhary JS, Grant SG. Comparative study of human and mouse postsynaptic proteomes finds high compositional conservation and abundance differences for key synaptic proteins. PLoS One. 2012;7((10)):e46683. doi: 10.1371/journal.pone.0046683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Citri A, Malenka RC. Synaptic plasticity: multiple forms, functions, and mechanisms. Neuropsychopharmacology. 2008 Jan;33((1)):18–41. doi: 10.1038/sj.npp.1301559. [DOI] [PubMed] [Google Scholar]
- 10.Collins MO, Husi H, Yu L, Brandon JM, Anderson CN, Blackstock WP, et al. Molecular characterization and comparison of the components and multiprotein complexes in the postsynaptic proteome. J Neurochem. 2006 Apr;97((Suppl 1)):16–23. doi: 10.1111/j.1471-4159.2005.03507.x. [DOI] [PubMed] [Google Scholar]
- 11.Fernández E, Collins MO, Uren RT, Kopanitsa MV, Komiyama NH, Croning MD, et al. Targeted tandem affinity purification of PSD-95 recovers core postsynaptic complexes and schizophrenia susceptibility proteins. Mol Syst Biol. 2009;5:269. doi: 10.1038/msb.2009.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schenck A, Bardoni B, Moro A, Bagni C, Mandel JL. A highly conserved protein family interacting with the fragile X mental retardation protein (FMRP) and displaying selective interactions with FMRP-related proteins FXR1P and FXR2P. Proc Natl Acad Sci USA. 2001;98((15)):8844–9. doi: 10.1073/pnas.151231598. Available from: http://www.ncbi.nlm.nih.gov/pubmed/11438699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Garber KB, Visootsak J, Warren ST. Fragile X syndrome. Eur J Hum Genet. 2008;16((6)):666–72. doi: 10.1038/ejhg.2008.61. Available from: http://www.ncbi.nlm.nih.gov/pubmed/18398441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Willemsen R, Bontekoe C, Tamanini F, Galjaard H, Hoogeveen A, Oostra B. Association of FMRP with ribosomal precursor particles in the nucleolus. Biochem Biophys Res Commun. 1996 Aug;225((1)):27–33. doi: 10.1006/bbrc.1996.1126. [DOI] [PubMed] [Google Scholar]
- 15.Christie SB, Akins MR, Schwob JE, Fallon JR. The FXG: a presynaptic fragile X granule expressed in a subset of developing brain circuits. J Neurosci. 2009;29((5)):1514–24. doi: 10.1523/JNEUROSCI.3937-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zorio DA, Jackson CM, Liu Y, Rubel EW, Wang Y. Cellular distribution of the fragile X mental retardation protein in the mouse brain. J Comp Neurol. 2017 Mar;525((4)):818–49. doi: 10.1002/cne.24100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Till SM, Wijetunge LS, Seidel VG, Harlow E, Wright AK, Bagni C, et al. Altered maturation of the primary somatosensory cortex in a mouse model of fragile X syndrome. Hum Mol Genet. 2012 May;21((10)):2143–56. doi: 10.1093/hmg/dds030. [DOI] [PubMed] [Google Scholar]
- 18.Siomi H, Siomi MC, Nussbaum RL, Dreyfuss G. The protein product of the fragile X gene, FMR1, has characteristics of an RNA-binding protein. Cell. 1993 Jul;74((2)):291–8. doi: 10.1016/0092-8674(93)90420-u. [DOI] [PubMed] [Google Scholar]
- 19.Darnell JC, Van Driesche SJ, Zhang C, Hung KY, Mele A, Fraser CE, et al. FMRP stalls ribosomal translocation on mRNAs linked to synaptic function and autism. Cell. 2011;146((2)):247–61. doi: 10.1016/j.cell.2011.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pasciuto E, Bagni C. SnapShot: FMRP mRNA targets and diseases. Cell. 2014 Sep;158((6)):1446–46.e1. doi: 10.1016/j.cell.2014.08.035. [DOI] [PubMed] [Google Scholar]
- 21.Zalfa F, Adinolfi S, Napoli I, Kühn-Hölsken E, Urlaub H, Achsel T, et al. Fragile X mental retardation protein (FMRP) binds specifically to the brain cytoplasmic RNAs BC1/BC200 via a novel RNA-binding motif. J Biol Chem. 2005;280((39)):33403–10. doi: 10.1074/jbc.M504286200. Available from: http://www.ncbi.nlm.nih.gov/pubmed/16006558. [DOI] [PubMed] [Google Scholar]
- 22.Zalfa F, Eleuteri B, Dickson KS, Mercaldo V, De Rubeis S, di Penta A, et al. A new function for the fragile X mental retardation protein in regulation of PSD-95 mRNA stability. Nat Neurosci. 2007;10((5)):578–87. doi: 10.1038/nn1893. Available from: http://www.ncbi.nlm.nih.gov/pubmed/17417632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.De Rubeis S, Bagni C. Regulation of molecular pathways in the fragile X syndrome: insights into autism spectrum disorders. J Neurodev Disord. 2011;3((3)):257–69. doi: 10.1007/s11689-011-9087-2. Available from: http://www.ncbi.nlm.nih.gov/pubmed/21842222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Darnell JC, Klann E. The translation of translational control by FMRP: therapeutic targets for FXS. Nat Neurosci. 2013;16((11)):1530–6. doi: 10.1038/nn.3379. Available from: http://www.ncbi.nlm.nih.gov/pubmed/23584741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Richter JD, Bassell GJ, Klann E. Dysregulation and restoration of translational homeostasis in fragile X syndrome. Nat Rev Neurosci. 2015 Oct;16((10)):595–605. doi: 10.1038/nrn4001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li Z, Zhang Y, Ku L, Wilkinson KD, Warren ST, Feng Y. The fragile X mental retardation protein inhibits translation via interacting with mRNA. Nucleic Acids Res. 2001;29((11)):2276–83. doi: 10.1093/nar/29.11.2276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Feng Y, Zhang F, Lokey LK, Chastain JL, Lakkis L, Eberhart D, et al. Translational suppression by trinucleotide repeat expansion at FMR1. Science. 1995 May;268((5211)):731–4. doi: 10.1126/science.7732383. [DOI] [PubMed] [Google Scholar]
- 28.Bramham CR, Wells DG. Dendritic mRNA: transport, translation and function. Nat Rev Neurosci. 2007 Oct;8((10)):776–89. doi: 10.1038/nrn2150. [DOI] [PubMed] [Google Scholar]
- 29.Antar LN, Afroz R, Dictenberg JB, Carroll RC, Bassell GJ. Metabotropic glutamate receptor activation regulates fragile X mental retardation protein and Fmr1 mRNA localization differentially in dendrites and at synapses. J Neurosci. 2004;24((11)):2648–55. doi: 10.1523/JNEUROSCI.0099-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bagni C, Greenough WT. From mRNP trafficking to spine dysmorphogenesis: the roots of fragile X syndrome. Nat Rev Neurosci. 2005 May;6((5)):376–87. doi: 10.1038/nrn1667. [DOI] [PubMed] [Google Scholar]
- 31.Napoli I, Mercaldo V, Boyl PP, Eleuteri B, Zalfa F, De Rubeis S, et al. The fragile X syndrome protein represses activity-dependent translation through CYFIP1, a new 4E-BP. Cell. 2008;134((6)):1042–54. doi: 10.1016/j.cell.2008.07.031. [DOI] [PubMed] [Google Scholar]
- 32.Bagni C, Zukin RS. A synaptic perspective of fragile X syndrome and autism spectrum disorders. Neuron. 2019 Mar;101((6)):1070–88. doi: 10.1016/j.neuron.2019.02.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hou L, Antion MD, Hu D, Spencer CM, Paylor R, Klann E. Dynamic translational and proteasomal regulation of fragile X mental retardation protein controls mGluR-dependent long-term depression. Neuron. 2006;51((4)):441–54. doi: 10.1016/j.neuron.2006.07.005. Available from: http://www.ncbi.nlm.nih.gov/pubmed/16908410. [DOI] [PubMed] [Google Scholar]
- 34.Nalavadi VC, Muddashetty RS, Gross C, Bassell GJ. Dephosphorylation-induced ubiquitination and degradation of FMRP in dendrites: a role in immediate early mGluR-stimulated translation. J Neurosci. 2012 Feb;32((8)):2582–7. doi: 10.1523/JNEUROSCI.5057-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Khayachi A, Gwizdek C, Poupon G, Alcor D, Chafai M, Cassé F, et al. Sumoylation regulates FMRP-mediated dendritic spine elimination and maturation. Nat Commun. 2018 Feb;9((1)):757. doi: 10.1038/s41467-018-03222-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Achsel T, Bagni C. Cooperativity in RNA-protein interactions: the complex is more than the sum of its partners. Curr Opin Neurobiol. 2016 Aug;39:146–51. doi: 10.1016/j.conb.2016.06.007. [DOI] [PubMed] [Google Scholar]
- 37.Banerjee A, Ifrim MF, Valdez AN, Raj N, Bassell GJ. Aberrant RNA translation in fragile X syndrome: from FMRP mechanisms to emerging therapeutic strategies. Brain Res. 2018 Aug;1693((Pt A)):24–36. doi: 10.1016/j.brainres.2018.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ifrim MF, Williams KR, Bassell GJ. Single-molecule imaging of PSD-95 mRNA translation in dendrites and its dysregulation in a mouse model of fragile X syndrome. J Neurosci. 2015 May;35((18)):7116–30. doi: 10.1523/JNEUROSCI.2802-14.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Davis JK, Broadie K. Multifarious functions of the fragile X mental retardation protein. Trends Genet. 2017;33((10)):703–14. doi: 10.1016/j.tig.2017.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Filippini A, Bonini D, Lacoux C, Pacini L, Zingariello M, Sancillo L, et al. Absence of the fragile X mental retardation protein results in defects of RNA editing of neuronal mRNAs in mouse. RNA Biol. 2017 Nov;14((11)):1–12. doi: 10.1080/15476286.2017.1338232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.De Rubeis S, Bagni C. Fragile X mental retardation protein control of neuronal mRNA metabolism: insights into mRNA stability. Mol Cell Neurosci. 2010;43((1)):43–50. doi: 10.1016/j.mcn.2009.09.013. Available from: http://www.ncbi.nlm.nih.gov/pubmed/19837168. [DOI] [PubMed] [Google Scholar]
- 42.Brown MR, Kronengold J, Gazula VR, Chen Y, Strumbos JG, Sigworth FJ, et al. Fragile X mental retardation protein controls gating of the sodium-activated potassium channel Slack. Nat Neurosci. 2010 Jul;13((7)):819–21. doi: 10.1038/nn.2563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Deng PY, Rotman Z, Blundon JA, Cho Y, Cui J, Cavalli V, et al. FMRP regulates neurotransmitter release and synaptic information transmission by modulating action potential duration via BK channels. Neuron. 2013;77((4)):696–711. doi: 10.1016/j.neuron.2012.12.018. Available from: http://www.ncbi.nlm.nih.gov/pubmed/23439122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Doll CA, Broadie K. Neuron class-specific requirements for fragile X mental retardation protein in critical period development of calcium signaling in learning and memory circuitry. Neurobiol Dis. 2016 May;89:76–87. doi: 10.1016/j.nbd.2016.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Doll CA, Broadie K. Activity-dependent FMRP requirements in development of the neural circuitry of learning and memory. Development. 2015;142((7)):1346–56. doi: 10.1242/dev.117127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hays SA, Huber KM, Gibson JR. Altered neocortical rhythmic activity states in Fmr1 KO mice are due to enhanced mGluR5 signaling and involve changes in excitatory circuitry. J Neurosci. 2011;31((40)):14223–1434. doi: 10.1523/JNEUROSCI.3157-11.2011. Available from: http://www.ncbi.nlm.nih.gov/pubmed/21976507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sabanov V, Braat S, D'Andrea L, Willemsen R, Zeidler S, Rooms L, et al. Impaired GABAergic inhibition in the hippocampus of Fmr1 knockout mice. Neuropharmacology. 2017 Apr;116:71–81. doi: 10.1016/j.neuropharm.2016.12.010. [DOI] [PubMed] [Google Scholar]
- 48.Maurin T, Bardoni B. Fragile X mental retardation protein: to Be or not to be a translational enhancer. Front Mol Biosci. 2018;5:113. doi: 10.3389/fmolb.2018.00113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Braat S, D'Hulst C, Heulens I, de Rubeis S, Mientjes E, Nelson DL, et al. The GABAA receptor is an FMRP target with therapeutic potential in fragile X syndrome. Cell Cycle. 2015;14((18)):2985–95. doi: 10.4161/15384101.2014.989114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Brown V, Jin P, Ceman S, Darnell JC, O'Donnell WT, Tenenbaum S, et al. Microarray identification of FMRP-associated brain mRNAs and altered mRNA translational profiles in fragile X syndrome. Cell. 2001;107((4)):477–87. doi: 10.1016/s0092-8674(01)00568-2. [DOI] [PubMed] [Google Scholar]
- 51.Ascano M, Mukherjee N, Bandaru P, Miller JB, Nusbaum JD, Corcoran DL, et al. FMRP targets distinct mRNA sequence elements to regulate protein expression. Nature. 2012;492((7429)):382–6. doi: 10.1038/nature11737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Maurin T, Lebrigand K, Castagnola S, Paquet A, Jarjat M, Popa A, et al. HITS-CLIP in various brain areas reveals new targets and new modalities of RNA binding by fragile X mental retardation protein. Nucleic Acids Res. 2018;46((12)):6344–55. doi: 10.1093/nar/gky267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Suhl J, Chopra P, Anderson BR, Bassell GJ, Warren ST. Analysis of FMRP mRNA target datasets reveals highly associated mRNAs mediated by G-quadruplex structures formed via clustered WGGA sequences. Hum Mol Genet. 2014;23((20)):5479–91. doi: 10.1093/hmg/ddu272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tabet R, Moutin E, Becker JA, Heintz D, Fouillen L, Flatter E, et al. Fragile X mental retardation protein (FMRP) controls diacylglycerol kinase activity in neurons. Proc Natl Acad Sci USA. 2016;113((26)):E3619–28. doi: 10.1073/pnas.1522631113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wang H, Wu LJ, Kim SS, Lee FJ, Gong B, Toyoda H, et al. FMRP acts as a key messenger for dopamine modulation in the forebrain. Neuron. 2008;59((4)):634–47. doi: 10.1016/j.neuron.2008.06.027. [DOI] [PubMed] [Google Scholar]
- 56.Zalfa F, Giorgi M, Primerano B, Moro A, Di Penta A, Reis S, et al. The fragile X syndrome protein FMRP associates with BC1 RNA and regulates the translation of specific mRNAs at synapses. Cell. 2003;112((3)):317–27. doi: 10.1016/s0092-8674(03)00079-5. [DOI] [PubMed] [Google Scholar]
- 57.Niere F, Wilkerson JR, Huber KM. Evidence for a fragile X mental retardation protein-mediated translational switch in metabotropic glutamate receptor-triggered Arc translation and long-term depression. J Neurosci. 2012;32((17)):5924–36. doi: 10.1523/JNEUROSCI.4650-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Clifton N, Rees E, Holmans P, Pardinas A, Harwood J, Florio A, et al. Genetic association of FMRP targets with psychiatric disorders. bioRxiv. 2020;2020 doi: 10.1038/s41380-020-00912-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sidorov MS, Auerbach BD, Bear MF. Fragile X mental retardation protein and synaptic plasticity. Mol Brain. 2013;6:15. doi: 10.1186/1756-6606-6-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bureau I, Shepherd GM, Svoboda K. Circuit and plasticity defects in the developing somatosensory cortex of Fmr1 knock-out mice. J Neurosci. 2008;28((20)):5178–88. doi: 10.1523/JNEUROSCI.1076-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hanson JE, Madison DV. Presynaptic Fmr1 genotype influences the degree of synaptic connectivity in a mosaic mouse model of fragile X syndrome. J Neurosci. 2007;27((15)):4014–8. doi: 10.1523/JNEUROSCI.4717-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Pan L, Zhang YQ, Woodruff E, Broadie K. The Drosophila fragile X gene negatively regulates neuronal elaboration and synaptic differentiation. Curr Biol. 2004;14((20)):1863–70. doi: 10.1016/j.cub.2004.09.085. [DOI] [PubMed] [Google Scholar]
- 63.Abekhoukh S, Bardoni B. CYFIP family proteins between autism and intellectual disability: links with fragile X syndrome. Front Cell Neurosci. 2014;8:81. doi: 10.3389/fncel.2014.00081. Available from: http://www.ncbi.nlm.nih.gov/pubmed/24733999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kobayashi K, Kuroda S, Fukata M, Nakamura T, Nagase T, Nomura N, et al. p140Sra-1 (specifically Rac1-associated protein) is a novel specific target for Rac1 small GTPase. J Biol Chem. 1998;273((1)):291–5. doi: 10.1074/jbc.273.1.291. Available from: http://www.ncbi.nlm.nih.gov/pubmed/9417078. [DOI] [PubMed] [Google Scholar]
- 65.Schenck A, Bardoni B, Langmann C, Harden N, Mandel JL, Giangrande A. CYFIP/Sra-1 controls neuronal connectivity in Drosophila and links the Rac1 GTPase pathway to the fragile X protein. Neuron. 2003;38((6)):887–98. doi: 10.1016/s0896-6273(03)00354-4. Available from: http://www.ncbi.nlm.nih.gov/pubmed/12818175. [DOI] [PubMed] [Google Scholar]
- 66.Richter JD, Sonenberg N. Regulation of cap-dependent translation by eIF4E inhibitory proteins. Nature. 2005;433((7025)):477–80. doi: 10.1038/nature03205. Available from: http://www.ncbi.nlm.nih.gov/pubmed/15690031. [DOI] [PubMed] [Google Scholar]
- 67.Marcotrigiano J, Gingras AC, Sonenberg N, Burley SK. Cap-dependent translation initiation in eukaryotes is regulated by a molecular mimic of eIF4G. Mol Cell. 1999;3((6)):707–16. doi: 10.1016/s1097-2765(01)80003-4. Available from: http://www.ncbi.nlm.nih.gov/pubmed/10394359. [DOI] [PubMed] [Google Scholar]
- 68.Panja D, Kenney JW, D'Andrea L, Zalfa F, Vedeler A, Wibrand K, et al. Two-stage translational control of dentate gyrus LTP consolidation is mediated by sustained BDNF-TrkB signaling to MNK. Cell Rep. 2014;9((4)):1430–45. doi: 10.1016/j.celrep.2014.10.016. Available from: https://www.ncbi.nlm.nih.gov/pubmed/25453757. [DOI] [PubMed] [Google Scholar]
- 69.Takenawa T, Suetsugu S. The WASP-WAVE protein network: connecting the membrane to the cytoskeleton. Nat Rev Mol Cell Biol. 2007;8((1)):37–48. doi: 10.1038/nrm2069. Available from: http://www.ncbi.nlm.nih.gov/pubmed/17183359. [DOI] [PubMed] [Google Scholar]
- 70.Eden S, Rohatgi R, Podtelejnikov AV, Mann M, Kirschner MW. Mechanism of regulation of WAVE1-induced actin nucleation by Rac1 and Nck. Nature. 2002;418((6899)):790–3. doi: 10.1038/nature00859. Available from: http://www.ncbi.nlm.nih.gov/pubmed/12181570. [DOI] [PubMed] [Google Scholar]
- 71.Chen Z, Borek D, Padrick SB, Gomez TS, Metlagel Z, Ismail AM, et al. Structure and control of the actin regulatory WAVE complex. Nature. 2010;468((7323)):533–8. doi: 10.1038/nature09623. Available from: http://www.ncbi.nlm.nih.gov/pubmed/21107423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kim Y, Sung JY, Ceglia I, Lee KW, Ahn JH, Halford JM, et al. Phosphorylation of WAVE1 regulates actin polymerization and dendritic spine morphology. Nature. 2006;442((7104)):814–7. doi: 10.1038/nature04976. Available from: http://www.ncbi.nlm.nih.gov/pubmed/16862120. [DOI] [PubMed] [Google Scholar]
- 73.Yan Z, Kim E, Datta D, Lewis DA, Soderling SH. Synaptic actin dysregulation, a convergent mechanism of mental disorders? J Neurosci. 2016 Nov;36((45)):11411–7. doi: 10.1523/JNEUROSCI.2360-16.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.De Rubeis S, Pasciuto E, Li KW, Fernández E, Di Marino D, Buzzi A, et al. CYFIP1 coordinates mRNA translation and cytoskeleton remodeling to ensure proper dendritic Spine formation. Neuron. 2013;79((6)):1169–82. doi: 10.1016/j.neuron.2013.06.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Di Marino D, D'Annessa I, Tancredi H, Bagni C, Gallicchio E. A unique binding mode of the eukaryotic translation initiation factor 4E for guiding the design of novel peptide inhibitors. Protein Sci. 2015 Sep;24((9)):1370–82. doi: 10.1002/pro.2708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Klann E, Dever TE. Biochemical mechanisms for translational regulation in synaptic plasticity. Nat Rev Neurosci. 2004 Dec;5((12)):931–42. doi: 10.1038/nrn1557. [DOI] [PubMed] [Google Scholar]
- 77.Costa-Mattioli M, Sossin WS, Klann E, Sonenberg N. Translational control of long-lasting synaptic plasticity and memory. Neuron. 2009 Jan;61((1)):10–26. doi: 10.1016/j.neuron.2008.10.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Gal-Ben-Ari S, Kenney JW, Ounalla-Saad H, Taha E, David O, Levitan D, et al. Consolidation and translation regulation. Learn Mem. 2012;19((9)):410–22. doi: 10.1101/lm.026849.112. Available from: http://www.ncbi.nlm.nih.gov/pubmed/22904372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Hsiao K, Harony-Hsiao K, Harony-Nicolas H, Buxbaum JD, Bozdagi-Gunal O, Benson DL. Cyfip1 regulates presynaptic activity during development. J Neurosci. 2016;36((5)):1564–76. doi: 10.1523/JNEUROSCI.0511-15.2016. Available from: http://www.ncbi.nlm.nih.gov/pubmed/26843638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Zhao L, Wang D, Wang Q, Rodal AA, Zhang YQ. Drosophila cyfip regulates synaptic development and endocytosis by suppressing filamentous actin assembly. Plos Genet. 2013 Apr;9((4)):e1003450. doi: 10.1371/journal.pgen.1003450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Domínguez-Iturza N, Lo AC, Shah D, Armendáriz M, Vannelli A, Mercaldo V, et al. The autism- and schizophrenia-associated protein CYFIP1 regulates bilateral brain connectivity and behaviour. Nat Commun. 2019 Aug;10((1)):3454. doi: 10.1038/s41467-019-11203-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Pathania M, Davenport EC, Muir J, Sheehan DF, López-Doménech G, Kittler JT. The autism and schizophrenia associated gene CYFIP1 is critical for the maintenance of dendritic complexity and the stabilization of mature spines. Transl Psychiatry. 2014;4:e374. doi: 10.1038/tp.2014.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Davenport EC, Szulc BR, Drew J, Taylor J, Morgan T, Higgs NF, et al. Autism and schizophrenia-associated CYFIP1 regulates the balance of synaptic excitation and inhibition. Cell Rep. 2019;26((8)):2037–e6. doi: 10.1016/j.celrep.2019.01.092. Available from: http://www.ncbi.nlm.nih.gov/pubmed/30784587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Bozdagi O, Sakurai T, Dorr N, Pilorge M, Takahashi N, Buxbaum JD. Haploinsufficiency of Cyfip1 produces fragile X-like phenotypes in mice. PLoS One. 2012;7((8)):e42422. doi: 10.1371/journal.pone.0042422. Available from: http://www.ncbi.nlm.nih.gov/pubmed/22900020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Föcking M, Lopez LM, English JA, Dicker P, Wolff A, Brindley E, et al. Proteomic and genomic evidence implicates the postsynaptic density in schizophrenia. Mol Psychiatry. 2015;20((4)):424–32. doi: 10.1038/mp.2014.63. [DOI] [PubMed] [Google Scholar]
- 86.Ehninger D, Silva AJ. Genetics and neuropsychiatric disorders: treatment during adulthood. Nat Med. 2009;15((8)):849–50. doi: 10.1038/nm0809-849. Available from: http://www.ncbi.nlm.nih.gov/pubmed/19661989. [DOI] [PubMed] [Google Scholar]
- 87.Melom JE, Littleton JT. Synapse development in health and disease. Curr Opin Genet Dev. 2011;21((3)):256–61. doi: 10.1016/j.gde.2011.01.002. Available from: http://www.ncbi.nlm.nih.gov/pubmed/21277192. [DOI] [PubMed] [Google Scholar]
- 88.Littleton SGN. Synaptopathies: diseases of the synaptome. Curr Opin Neurobiol. 2012 Jun;22((3)):522–9. doi: 10.1016/j.conb.2012.02.002. [DOI] [PubMed] [Google Scholar]
- 89.Owen MJ, O'Donovan MC, Harrison PJ. Schizophrenia: a genetic disorder of the synapse? BMJ. 2005 Jan;330((7484)):158–9. doi: 10.1136/bmj.330.7484.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Guang S, Pang N, Deng X, Yang L, He F, Wu L, et al. Synaptopathology involved in autism spectrum disorder. Front Cell Neurosci. 2018;12:470. doi: 10.3389/fncel.2018.00470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Hall J, Trent S, Thomas KL, O'Donovan MC, Owen MJ. Genetic risk for schizophrenia: convergence on synaptic pathways involved in plasticity. Biol Psychiatry. 2015;77((1)):52–8. doi: 10.1016/j.biopsych.2014.07.011. [DOI] [PubMed] [Google Scholar]
- 92.Kirov G, Pocklington AJ, Holmans P, Ivanov D, Ikeda M, Ruderfer D, et al. De novo CNV analysis implicates specific abnormalities of postsynaptic signalling complexes in the pathogenesis of schizophrenia. Mol Psychiatry. 2012 Feb;17((2)):142–53. doi: 10.1038/mp.2011.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Guilmatre A, Dubourg C, Mosca AL, Legallic S, Goldenberg A, Drouin-Garraud V, et al. Recurrent rearrangements in synaptic and neurodevelopmental genes and shared biologic pathways in schizophrenia, autism, and mental retardation. Arch Gen Psychiatry. 2009;66((9)):947–56. doi: 10.1001/archgenpsychiatry.2009.80. Available from: http://www.ncbi.nlm.nih.gov/pubmed/19736351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Pocklington AJ, O'Donovan M, Owen MJ. The synapse in schizophrenia. Eur J Neurosci. 2014;39((7)):1059–67. doi: 10.1111/ejn.12489. [DOI] [PubMed] [Google Scholar]
- 95.Kaizuka T, Takumi T. Postsynaptic density proteins and their involvement in neurodevelopmental disorders. J Biochem. 2018 Jun;163((6)):447–55. doi: 10.1093/jb/mvy022. [DOI] [PubMed] [Google Scholar]
- 96.Buffington SA, Huang W, Costa-Mattioli M. Translational control in synaptic plasticity and cognitive dysfunction. Annu Rev Neurosci. 2014;37:17–38. doi: 10.1146/annurev-neuro-071013-014100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Santini E, Klann E. Reciprocal signaling between translational control pathways and synaptic proteins in autism spectrum disorders. Sci Signal. 2014 Oct;7((349)):re10. doi: 10.1126/scisignal.2005832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Amorim IS, Lach G, Gkogkas CG. The role of the eukaryotic translation initiation factor 4E (eIF4E) in neuropsychiatric disorders. Front Genet. 2018;9:561. doi: 10.3389/fgene.2018.00561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Horváth S, Mirnics K. Schizophrenia as a disorder of molecular pathways. Biol Psychiatry. 2015 Jan;77((1)):22–8. doi: 10.1016/j.biopsych.2014.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Abrahams BS, Geschwind DH. Advances in autism genetics: on the threshold of a new neurobiology. Nat Rev Genet. 2008 May;9((5)):341–55. doi: 10.1038/nrg2346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Subramanian M, Timmerman CK, Schwartz JL, Pham DL, Meffert MK. Characterizing autism spectrum disorders by key biochemical pathways. Front Neurosci. 2015;9:313. doi: 10.3389/fnins.2015.00313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Forrest MP, Parnell E, Penzes P. Dendritic structural plasticity and neuropsychiatric disease. Nat Rev Neurosci. 2018 Mar;19((4)):215–34. doi: 10.1038/nrn.2018.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Catterall WA, Leal K, Nanou E. Calcium channels and short-term synaptic plasticity. J Biol Chem. 2018;288((15)):10742–9. doi: 10.1074/jbc.R112.411645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Bourgeron T. From the genetic architecture to synaptic plasticity in autism spectrum disorder. Nat Rev Neurosci. 2015;16((9)):551–63. doi: 10.1038/nrn3992. [DOI] [PubMed] [Google Scholar]
- 105.Averbeck BB, Chafee MV. Using model systems to understand errant plasticity mechanisms in psychiatric disorders. Nat Neurosci. 2016 Nov;19((11)):1418–25. doi: 10.1038/nn.4413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Shepherd GM. Corticostriatal connectivity and its role in disease. Nat Rev Neurosci. 2013;14((4)):278–91. doi: 10.1038/nrn3469. Available from: http://www.ncbi.nlm.nih.gov/pubmed/23511908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Owen MJ, Sawa A, Mortensen PB. Schizophrenia. Lancet. 2016;6736((15)):1–12. doi: 10.1016/S0140-6736(15)01121-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Jacquemont S, Hagerman RJ, Hagerman PJ, Leehey MA. Fragile-X syndrome and fragile X-associated tremor/ataxia syndrome: two faces of FMR1. Lancet Neurol. 2007;6((1)):45–5. doi: 10.1016/S1474-4422(06)70676-7. Available from: http://www.ncbi.nlm.nih.gov/pubmed/17166801. [DOI] [PubMed] [Google Scholar]
- 109.Santoro MR, Bray SM, Warren ST. Molecular mechanisms of fragile X syndrome: a twenty-year perspective. Annu Rev Pathol. 2012;7((1)):219–45. doi: 10.1146/annurev-pathol-011811-132457. [DOI] [PubMed] [Google Scholar]
- 110.Irwin SA, Patel B, Idupulapati M, Harris JB, Crisostomo RA, Larsen BP, et al. Abnormal dendritic spine characteristics in the temporal and visual cortices of patients with fragile-X syndrome: a quantitative examination. Am J Med Genet. 2001;98((2)):161–7. doi: 10.1002/1096-8628(20010115)98:2<161::aid-ajmg1025>3.0.co;2-b. Available from: http://www.ncbi.nlm.nih.gov/pubmed/11223852. [DOI] [PubMed] [Google Scholar]
- 111.Hinton VJ, Brown WT, Wisniewski K, Rudelli RD. Analysis of neocortex in three males with the fragile X syndrome. Am J Med Genet. 1991 Dec;41((3)):289–94. doi: 10.1002/ajmg.1320410306. [DOI] [PubMed] [Google Scholar]
- 112.Martinez-Cerdeno V. Dendrite and spine modifications in autism and related neurodevelopmental disorders in patients and animal models. Dev Neurobiol. 2017 Apr;77((4)):393–404. doi: 10.1002/dneu.22417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Jacquemont S, Pacini L, Jønch AE, Cencelli G, Rozenberg I, He Y, et al. Protein synthesis levels are increased in a subset of individuals with fragile X syndrome. Hum Mol Genet. 2018 Jun;27((21)):3825–51. doi: 10.1093/hmg/ddy291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Qin M, Schmidt KC, Zametkin AJ, Bishu S, Horowitz LM, Burlin TV, et al. Altered cerebral protein synthesis in fragile X syndrome: studies in human subjects and knockout mice. J Cereb Blood Flow Metab. 2013;33((4)):499–507. doi: 10.1038/jcbfm.2012.205. Available from: http://www.ncbi.nlm.nih.gov/pubmed/23299245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Contractor A, Klyachko VA, Portera-Cailliau C. Altered neuronal and circuit excitability in fragile X syndrome. Neuron. 2015 Aug;87((4)):699–715. doi: 10.1016/j.neuron.2015.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Swanson MR, Wolff JJ, Shen MD, Styner M, Estes A, Gerig G, et al. Development of white matter circuitry in infants with fragile X syndrome. JAMA Psychiatry. 2018 May;75((5)):505–13. doi: 10.1001/jamapsychiatry.2018.0180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Belmonte MK, Bourgeron T. Fragile X syndrome and autism at the intersection of genetic and neural networks. Nat Neurosci. 2006;9((10)):1221–5. doi: 10.1038/nn1765. Available from: http://www.ncbi.nlm.nih.gov/pubmed/17001341. [DOI] [PubMed] [Google Scholar]
- 118.Turk J. Fragile X syndrome: lifespan developmental implications for those without as well as with intellectual disability. Curr Opin Psychiatry. 2011;24((5)):387–97. doi: 10.1097/YCO.0b013e328349bb77. Available from: http://www.ncbi.nlm.nih.gov/pubmed/21825875. [DOI] [PubMed] [Google Scholar]
- 119.Bagni C, Tassone F, Neri G, Hagerman R. Fragile X syndrome: causes, diagnosis, mechanisms, and therapeutics. J Clin Inves. 2012;122((12)):4314–22. doi: 10.1172/JCI63141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Auerbach BD, Osterweil EK, Bear MF. Mutations causing syndromic autism define an axis of synaptic pathophysiology. Nature. 2011;480((7375)):63–8. doi: 10.1038/nature10658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Fatemi SH, Kneeland RE, Liesch SB, Folsom TD. Fragile X mental retardation protein levels are decreased in major psychiatric disorders. Vol. 124. Netherlands: Schizophrenia Research; 2010. pp. p. 246–247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Kovács T, Kelemen O, Kéri S. Decreased fragile X mental retardation protein (FMRP) is associated with lower IQ and earlier illness onset in patients with schizophrenia. Psychiatry Res. 2013 Dec;210((3)):690–3. doi: 10.1016/j.psychres.2012.12.022. [DOI] [PubMed] [Google Scholar]
- 123.Ashworth A, Abusaad I, Walsh C, Nanko S, Murray RM, Asherson P, et al. Linkage analysis of the fragile X gene FMR-1 and schizophrenia: no evidence for linkage but report of a family with schizophrenia and an unstable triplet repeat. Psychiatr Genet. 1996;6((2)):81–6. doi: 10.1097/00041444-199622000-00008. [DOI] [PubMed] [Google Scholar]
- 124.Jönsson E, Björck E, Wahlström J, Gustavsson P, Sedvall G. Screening for CGG trinucleotide repeat expansion in the fragile X mental retardation 1 gene in schizophrenic patients. Psychiatr Genet. 1995;5((4)):157–60. doi: 10.1097/00041444-199524000-00002. [DOI] [PubMed] [Google Scholar]
- 125.Khin NA, Tarleton J, Raghu B, Park SK. Clinical description of an adult male with psychosis who showed FMR1 gene methylation mosaicism. Am J Med Genet. 1998 May;81((3)):222–4. doi: 10.1002/(sici)1096-8628(19980508)81:3<222::aid-ajmg3>3.0.co;2-y. [DOI] [PubMed] [Google Scholar]
- 126.The Dutch-Belgian Fragile X Consortium Fmr1 knockout mice: a model to study fragile X mental retardation. The Dutch-Belgian fragile X consortium. Cell. 1994;78((1)):23–3. Available from: http://www.ncbi.nlm.nih.gov/pubmed/8033209. [PubMed] [Google Scholar]
- 127.Till SM, Asiminas A, Jackson AD, Katsanevaki D, Barnes SA, Osterweil EK, et al. Conserved hippocampal cellular pathophysiology but distinct behavioural deficits in a new rat model of FXS. Hum Mol Genet. 2015 Nov;24((21)):5977–84. doi: 10.1093/hmg/ddv299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Asiminas A, Jackson AD, Louros SR, Till SM, Spano T, Dando O, et al. Sustained correction of associative learning deficits after brief, early treatment in a rat model of fragile X syndrome. Sci Transl Med. 2019 May;11((494)):eaao0498. doi: 10.1126/scitranslmed.aao0498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Grossman AW, Elisseou NM, McKinney BC, Greenough WT. Hippocampal pyramidal cells in adult Fmr1 knockout mice exhibit an immature-appearing profile of dendritic spines. Brain Res. 2006;1084((1)):158–64. doi: 10.1016/j.brainres.2006.02.044. Available from: http://www.ncbi.nlm.nih.gov/pubmed/16574084. [DOI] [PubMed] [Google Scholar]
- 130.Wijetunge LS, Angibaud J, Frick A, Kind PC, Nägerl UV. Stimulated emission depletion (STED) microscopy reveals nanoscale defects in the developmental trajectory of dendritic spine morphogenesis in a mouse model of fragile X syndrome. J Neurosci. 2014 Apr;34((18)):6405–12. doi: 10.1523/JNEUROSCI.5302-13.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Qin M, Kang J, Burlin TV, Jiang C, Smith CB. Postadolescent changes in regional cerebral protein synthesis: an in vivo study in the FMR1 null mouse. J Neurosci. 2005 May;25((20)):5087–95. doi: 10.1523/JNEUROSCI.0093-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Dölen G, Osterweil E, Rao BS, Smith GB, Auerbach BD, Chattarji S, et al. Correction of fragile X syndrome in mice. Neuron. 2007;56((6)):955–62. doi: 10.1016/j.neuron.2007.12.001. Available from: http://www.ncbi.nlm.nih.gov/pubmed/18093519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Osterweil EK, Krueger DD, Reinhold K, Bear MF. Hypersensitivity to mGluR5 and ERK1/2 leads to excessive protein synthesis in the hippocampus of a mouse model of fragile X syndrome. J Neurosci. 2010 Nov;30((46)):15616–27. doi: 10.1523/JNEUROSCI.3888-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Bernardet M, Crusio WE. Fmr1 KO mice as a possible model of autistic features. ScientificWorldJournal. 2006;6:1164–76. doi: 10.1100/tsw.2006.220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Mineur YS, Huynh LX, Crusio WE. Social behavior deficits in the Fmr1 mutant mouse. Behav Brain Res. 2006;168((1)):172–5. doi: 10.1016/j.bbr.2005.11.004. Available from: http://www.ncbi.nlm.nih.gov/pubmed/16343653. [DOI] [PubMed] [Google Scholar]
- 136.Mines MA, Yuskaitis CJ, King MK, Beurel E, Jope RS. GSK3 influences social preference and anxiety-related behaviors during social interaction in a mouse model of fragile X syndrome and autism. PLoS One. 2010;5((3)):e9706. doi: 10.1371/journal.pone.0009706. Available from: http://www.ncbi.nlm.nih.gov/pubmed/20300527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Oddi D, Crusio WE, D'Amato FR, Pietropaolo S. Monogenic mouse models of social dysfunction: implications for autism. Behav Brain Res. 2013;251:75–84. doi: 10.1016/j.bbr.2013.01.002. Available from: http://www.ncbi.nlm.nih.gov/pubmed/23327738. [DOI] [PubMed] [Google Scholar]
- 138.Baker KB, Wray SP, Ritter R, Mason S, Lanthorn TH, Savelieva KV. Male and female Fmr1 knockout mice on C57 albino background exhibit spatial learning and memory impairments. Genes Brain Behav. 2010 Aug;9((6)):562–74. doi: 10.1111/j.1601-183X.2010.00585.x. [DOI] [PubMed] [Google Scholar]
- 139.Van Dam D, D'Hooge R, Hauben E, Reyniers E, Gantois I, Bakker CE, et al. Spatial learning, contextual fear conditioning and conditioned emotional response in Fmr1 knockout mice. Behav Brain Res. 2000;117((1–2)):127–36. doi: 10.1016/s0166-4328(00)00296-5. Available from: http://www.ncbi.nlm.nih.gov/pubmed/11099766. [DOI] [PubMed] [Google Scholar]
- 140.Paradee W, Melikian HE, Rasmussen DL, Kenneson A, Conn PJ, Warren ST. Fragile X mouse: strain effects of knockout phenotype and evidence suggesting deficient amygdala function. Neuroscience. 1999;94((1)):185–92. doi: 10.1016/s0306-4522(99)00285-7. Available from: http://www.ncbi.nlm.nih.gov/pubmed/10613508. [DOI] [PubMed] [Google Scholar]
- 141.D'Hooge R, Nagels G, Franck F, Bakker CE, Reyniers E, Storm K, et al. Mildly impaired water maze performance in male Fmr1 knockout mice. Neuroscience. 1997;76((2)):367–76. doi: 10.1016/s0306-4522(96)00224-2. Available from: http://www.ncbi.nlm.nih.gov/pubmed/9015322. [DOI] [PubMed] [Google Scholar]
- 142.Richter JD, Klann E. Making synaptic plasticity and memory last: mechanisms of translational regulation. Genes Dev. 2009;23((1)):1–11. doi: 10.1101/gad.1735809. Available from: http://www.ncbi.nlm.nih.gov/pubmed/19136621. [DOI] [PubMed] [Google Scholar]
- 143.Weiler IJ, Spangler CC, Klintsova AY, Grossman AW, Kim SH, Bertaina-Anglade V, et al. Fragile X mental retardation protein is necessary for neurotransmitter-activated protein translation at synapses. Proc Natl Acad Sci USA. 2004 Dec;101((50)):17504–9. doi: 10.1073/pnas.0407533101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Huber KM, Gallagher SM, Warren ST, Bear MF. Altered synaptic plasticity in a mouse model of fragile X mental retardation. Proc Natl Acad Sci USA. 2002;99((11)):7746–50. doi: 10.1073/pnas.122205699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Bear MF, Huber KM, Warren ST. The mGluR theory of fragile X mental retardation. Trends Neurosci. 2004;27((7)):370–7. doi: 10.1016/j.tins.2004.04.009. Available from: http://www.ncbi.nlm.nih.gov/pubmed/15219735. [DOI] [PubMed] [Google Scholar]
- 146.de Vrij FM, Levenga J, van der Linde HC, Koekkoek SK, De Zeeuw CI, Nelson DL, et al. Rescue of behavioral phenotype and neuronal protrusion morphology in Fmr1 KO mice. Neurobiol Dis. 2008;31((1)):127–32. doi: 10.1016/j.nbd.2008.04.002. Available from: http://www.ncbi.nlm.nih.gov/pubmed/18571098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Ronesi JA, Collins KA, Hays SA, Tsai NP, Guo W, Birnbaum SG, et al. Disrupted Homer scaffolds mediate abnormal mGluR5 function in a mouse model of fragile X syndrome. Nat Neurosci. 2012 Mar;15((3)):431–40, S1. doi: 10.1038/nn.3033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Clifton NE, Trent S, Thomas KL, Hall J. Regulation and function of activity-dependent homer in synaptic plasticity. Mol Neuropsychiatry. 2019 Jun;5((3)):147–61. doi: 10.1159/000500267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Sharma A, Hoeffer CA, Takayasu Y, Miyawaki T, McBride SM, Klann E, et al. Dysregulation of mTOR signaling in fragile X syndrome. J Neurosci. 2010 Jan;30((2)):694–702. doi: 10.1523/JNEUROSCI.3696-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Santini E, Huynh TN, Longo F, Koo SY, Mojica E, D'Andrea L, et al. Reducing eIF4E-eIF4G interactions restores the balance between protein synthesis and actin dynamics in fragile X syndrome model mice. Sci Signal. 2017 Nov;10((504)):eaan0665. doi: 10.1126/scisignal.aan0665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Pop AS, Gomez-Mancilla B, Neri G, Willemsen R, Gasparini F. Fragile X syndrome: a preclinical review on metabotropic glutamate receptor 5 (mGluR5) antagonists and drug development. Psychopharmacology. 2014 Mar;231((6)):1217–26. doi: 10.1007/s00213-013-3330-3. [DOI] [PubMed] [Google Scholar]
- 152.Castagnola S, Bardoni B, Maurin T. The search for an effective therapy to treat fragile X syndrome: dream or reality? Front Synaptic Neurosci. 2017;9:15. doi: 10.3389/fnsyn.2017.00015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Suhl JA, Warren ST. Single-nucleotide mutations in FMR1 reveal novel functions and regulatory mechanisms of the fragile X syndrome protein FMRP. J Exp Neurosci. 2015;9((Suppl 2)):35–41. doi: 10.4137/JEN.S25524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Handt M, Epplen A, Hoffjan S, Mese K, Epplen JT, Dekomien G. Point mutation frequency in the FMR1 gene as revealed by fragile X syndrome screening. Mol Cell Probes. 2014;28((5–6)):279–83. doi: 10.1016/j.mcp.2014.08.003. [DOI] [PubMed] [Google Scholar]
- 155.Grønskov K, Brøndum-Nielsen K, Dedic A, Hjalgrim H. A nonsense mutation in FMR1 causing fragile X syndrome. Eur J Hum Genet. 2011;19((4)):489–91. doi: 10.1038/ejhg.2010.223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Sitzmann AF, Hagelstrom RT, Tassone F, Hagerman RJ, Butler MG. Rare FMR1 gene mutations causing fragile X syndrome: a review. Am J Med Genet A. 2018;176((1)):11–8. doi: 10.1002/ajmg.a.38504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Collins SC, Bray SM, Suhl JA, Cutler DJ, Coffee B, Zwick ME, et al. Identification of novel FMR1 variants by massively parallel sequencing in developmentally delayed males. Am J Med Genet A. 2010;152((10)):2512–20. doi: 10.1002/ajmg.a.33626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Saré RM, Figueroa C, Lemons A, Loutaev I, Smith CB. Comparative behavioral phenotypes of fmr1 KO, fxr2 het, and fmr1 KO/fxr2 het mice. Brain Sci. 2019 Jan;9((1)):13. doi: 10.3390/brainsci9010013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Kuroda Y, Ohashi I, Tominaga M, Saito T, Nagai J, Ida K, et al. De novo duplication of 17p13.1–p13.2 in a patient with intellectual disability and obesity. Am J Med Genet A. 2014 Jun;164((6)):1550–4. doi: 10.1002/ajmg.a.36477. [DOI] [PubMed] [Google Scholar]
- 160.Giordano L, Palestra F, Giuffrida MG, Molinaro A, Iodice A, Bernardini L, et al. 17p13.17p13.1 microdeletion: genetic and clinical findings in a new patient with epilepsy and comparison with literature. Am J Med Genet A. 2014 Jan;164((1)):225–30. doi: 10.1002/ajmg.a.36225. [DOI] [PubMed] [Google Scholar]
- 161.Ripke S, Neale BM, Corvin A, Walters JTR, Farh K-H, Holmans P, et al. Biological insights from 108 schizophrenia-associated genetic loci. Nature. 2014;511((7510)):421–7. doi: 10.1038/nature13595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Pardiñas AF, Holmans P, Holmans P, Pocklington AJ, Escott-Price V, Ripke S, et al. Common schizophrenia alleles are enriched in mutation-intolerant genes and in regions under strong background selection. Nat Genet. 2018;50((3)):381–9. doi: 10.1038/s41588-018-0059-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Liu X, Kelsoe JR, Kelsoe JR, Greenwood TA, Green-Wood TA, Nievergelt CM, et al. A genome-wide association study of bipolar disorder with comorbid eating disorder replicates the SOX2-OT region. J Affect Disord. 2016;189:141–9. doi: 10.1016/j.jad.2015.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Takata A, Matsumoto N, Kato T. Genome-wide identification of splicing QTLs in the human brain and their enrichment among schizophrenia-associated loci. Nat Commun. 2017;8:14519. doi: 10.1038/ncomms14519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Stepniak B, Kästner A, Poggi G, Mitjans M, Begemann M, Hartmann A, et al. Accumulated common variants in the broader fragile X gene family modulate autistic phenotypes. EMBO Mol Med. 2015;7((12)):1565–79. doi: 10.15252/emmm.201505696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Jansen A, Dieleman GC, Smit AB, Verhage M, Verhulst FC, Polderman TJC, et al. Gene-set analysis shows association between FMRP targets and autism spectrum disorder. Eur J Hum Genet. 2017;25((7)):863–8. doi: 10.1038/ejhg.2017.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Wray NR, Ripke S, Mattheisen M, Trzaskowski M, Byrne EM, Abdellaoui A, et al. Genome-wide association analyses identify 44 risk variants and refine the genetic architecture of major depression. Nat Genet. 2018;50((5)):668–1. doi: 10.1038/s41588-018-0090-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Purcell SM, Moran JL, Fromer M, Ruderfer D, Solovieff N, Roussos P, et al. A polygenic burden of rare disruptive mutations in schizophrenia. Nature. 2014;506((7487)):185–90. doi: 10.1038/nature12975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Richards AL, Leonenko G, Walters JT, Kavanagh DH, Rees EG, Evans A, et al. Exome arrays capture polygenic rare variant contributions to schizophrenia. Hum Mol Genet. 2016;25((5)):1001–7. doi: 10.1093/hmg/ddv620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Leonenko G, Richards AL, Walters JT, Pocklington A, Chambert K, Al Eissa MM, et al. Mutation intolerant genes and targets of FMRP are enriched for nonsynonymous alleles in schizophrenia. Am J Med Genet B Neuropsychiatr Genet. 2017 Feb;174((7)):724–31. doi: 10.1002/ajmg.b.32560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Genovese G, Fromer M, Stahl EA, Ruderfer DM, Chambert K, Landén M, et al. Increased burden of ultra-rare protein-altering variants among 4,877 individuals with schizophrenia. Nat Neurosci. 2016;19((11)):1433–41. doi: 10.1038/nn.4402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.De Rubeis S, He X, Goldberg AP, Poultney CS, Samocha K, Cicek AE, et al. Synaptic, transcriptional and chromatin genes disrupted in autism. Nature. 2014;515((7526)):209–15. doi: 10.1038/nature13772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Goes FS, Pirooznia M, Parla JS, Kramer M, Ghiban E, Mavruk S, et al. Exome sequencing of familial bipolar disorder. JAMA Psychiatry. 2016;73((6)):590–7. doi: 10.1001/jamapsychiatry.2016.0251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Fromer M, Pocklington AJ, Kavanagh DH, Williams HJ, Dwyer S, Gormley P, et al. De novo mutations in schizophrenia implicate synaptic networks. Nature. 2014;506((7487)):179–84. doi: 10.1038/nature12929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Iossifov I, Ronemus M, Levy D, Wang Z, Hakker I, Rosenbaum J, et al. De novo gene disruptions in children on the autistic spectrum. Neuron. 2012;74((2)):285–99. doi: 10.1016/j.neuron.2012.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Iossifov I, O'Roak BJ, Sanders SJ, Ronemus M, Krumm N, Levy D, et al. The contribution of de novo coding mutations to autism spectrum disorder. Nature. 2014;515((7526)):216–21. doi: 10.1038/nature13908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Fernández E, Rajan N, Bagni C. The FMRP regulon: from targets to disease convergence. Front Neurosci. 2013;7:191. doi: 10.3389/fnins.2013.00191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Pocklington AJ, Rees E, Walters JT, Han J, Kavanagh DH, Chambert KD, et al. Novel findings from CNVs implicate inhibitory and excitatory signaling complexes in schizophrenia. Neuron. 2015;86((5)):1203–14. doi: 10.1016/j.neuron.2015.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Marshall CR, Howrigan DP, Merico D, Thiruvahindrapuram B, Wu W, Greer DS, et al. Contribution of copy number variants to schizophrenia from a genome-wide study of 41,321 subjects. Nat Genet. 2017;49((1)):27–35. doi: 10.1038/ng.3725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Szatkiewicz JP, O'Dushlaine C, Chen G, Chambert K, Moran JL, Neale BM, et al. Copy number variation in schizophrenia in Sweden. Mol Psychiatry. 2014;19((7)):762–73. doi: 10.1038/mp.2014.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Ouwenga RL, Dougherty J. Fmrp targets or not: long, highly brain-expressed genes tend to be implicated in autism and brain disorders. Mol Autism. 2015;6:16. doi: 10.1186/s13229-015-0008-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Cox DM, Butler MG. The 15q11.2 BP1–BP2 microdeletion syndrome: a review. Int J Mol Sci. 2015;16((2)):4068–82. doi: 10.3390/ijms16024068. Available from: http://www.ncbi.nlm.nih.gov/pubmed/25689425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Chai JH, Locke DP, Greally JM, Knoll JH, Ohta T, Dunai J, et al. Identification of four highly conserved genes between breakpoint hotspots BP1 and BP2 of the Prader-Willi/Angelman syndromes deletion region that have undergone evolutionary transposition mediated by flanking duplicons. Am J Hum Genet. 2003;73((4)):898–25. doi: 10.1086/378816. Available from: http://www.ncbi.nlm.nih.gov/pubmed/14508708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Butler MG, Bittel DC, Kibiryeva N, Talebizadeh Z, Thompson T. Behavioral differences among subjects with Prader-Willi syndrome and type I or type II deletion and maternal disomy. Pediatrics. 2004 Mar;113((3 Pt 1)):565–73. doi: 10.1542/peds.113.3.565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Bittel DC, Kibiryeva N, Butler MG. Expression of 4 genes between chromosome 15 breakpoints 1 and 2 and behavioral outcomes in Prader-Willi syndrome. Pediatrics. 2006;118((4)):e1276–83. doi: 10.1542/peds.2006-0424. Available from: http://www.ncbi.nlm.nih.gov/pubmed/16982806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Butler MG. Clinical and genetic aspects of the 15q11.2 BP1-BP2 microdeletion disorder. J Intellect Disabil Res. 2017;61((6)):568–79. doi: 10.1111/jir.12382. Available from: http://www.ncbi.nlm.nih.gov/pubmed/28387067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Burnside RD, Pasion R, Mikhail FM, Carroll AJ, Robin NH, Youngs EL, et al. Microdeletion/microduplication of proximal 15q11.2 between BP1 and BP2: a susceptibility region for neurological dysfunction including developmental and language delay. Hum Genet. 2011 Oct;130((4)):517–28. doi: 10.1007/s00439-011-0970-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Vanlerberghe C, Petit F, Malan V, Vincent-Delorme C, Bouquillon S, Boute O, et al. 15q11.2 microdeletion (BP1–BP2) and developmental delay, behaviour issues, epilepsy and congenital heart disease: a series of 52 patients. Eur J Med Genet. 2015 Mar;58((3)):140–7. doi: 10.1016/j.ejmg.2015.01.002. [DOI] [PubMed] [Google Scholar]
- 189.Woo YJ, Kanellopoulos AK, Hemati P, Kirschen J, Nebel RA, Wang T, et al. Domain-specific cognitive impairments in humans and flies with reduced CYFIP1 dosage. Biol Psychiatry. 2019 Aug;86((4)):306–14. doi: 10.1016/j.biopsych.2019.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Ulfarsson MO, Walters GB, Gustafsson O, Steinberg S, Silva A, Doyle OM, et al. 15q11.2 CNV affects cognitive, structural and functional correlates of dyslexia and dyscalculia. Transl Psychiatry. 2017 Apr;7((4)):e1109. doi: 10.1038/tp.2017.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Silva AI, Ulfarsson MO, Stefansson H, Gustafsson O, Walters GB, Linden DEJ, et al. Reciprocal white matter changes associated with copy number variation at 15q11.2 BP1–BP2: a diffusion tensor imaging study. Biol Psychiatry. 2019;85((7)):563–72. doi: 10.1016/j.biopsych.2018.11.004. Available from: https://www.ncbi.nlm.nih.gov/pubmed/30583851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Beaulieu C. The basis of anisotropic water diffusion in the nervous system: a technical review. NMR Biomed. 2002;15((7–8)):435–55. doi: 10.1002/nbm.782. [DOI] [PubMed] [Google Scholar]
- 193.Green T, Barnea-Goraly N, Raman M, Hall SS, Lightbody AA, Bruno JL, et al. Specific effect of the fragile-X mental retardation-1 gene (FMR1) on white matter microstructure. Br J Psychiatry. 2015 Aug;207((2)):143–8. doi: 10.1192/bjp.bp.114.151654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Cafferkey M, Ahn JW, Flinter F, Ogilvie C. Phenotypic features in patients with 15q11.2(BP1–BP2) deletion: further delineation of an emerging syndrome. Am J Med Genet A. 2014 Aug;164((8)):1916–22. doi: 10.1002/ajmg.a.36554. [DOI] [PubMed] [Google Scholar]
- 195.Stefansson H, Meyer-Lindenberg A, Steinberg S, Magnusdottir B, Morgen K, Arnarsdottir S, et al. CNVs conferring risk of autism or schizophrenia affect cognition in controls. Nature. 2014;505((7483)):361–6. doi: 10.1038/nature12818. Available from: https://www.ncbi.nlm.nih.gov/pubmed/24352232. [DOI] [PubMed] [Google Scholar]
- 196.Abekhoukh S, Sahin HB, Grossi M, Zongaro S, Maurin T, Madrigal I, et al. New insights into the regulatory function of CYFIP1 in the context of WAVE- and FMRP-containing complexes. Dis Model Mech. 2017;10((4)):463–74. doi: 10.1242/dmm.025809. Available from: http://www.ncbi.nlm.nih.gov/pubmed/28183735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Yoon KJ, Nguyen HN, Ursini G, Zhang F, Kim NS, Wen Z, et al. Modeling a genetic risk for schizophrenia in iPSCs and mice reveals neural stem cell deficits associated with adherens junctions and polarity. Cell Stem Cell. 2014;15((1)):79–91. doi: 10.1016/j.stem.2014.05.003. Available from: http://www.ncbi.nlm.nih.gov/pubmed/24996170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Nebel RA, Zhao D, Pedrosa E, Kirschen J, Lachman HM, Zheng D, et al. Reduced CYFIP1 in human neural progenitors results in dysregulation of schizophrenia and epilepsy gene networks. PLoS One. 2016;11((1)):e0148039. doi: 10.1371/journal.pone.0148039. Available from: http://www.ncbi.nlm.nih.gov/pubmed/26824476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Das DK, Tapias V, D'Aiuto L, Chowdari KV, Francis L, Zhi Y, et al. Genetic and morphological features of human iPSC-derived neurons with chromosome 15q11.2 (BP1–BP2) deletions. Mol Neuropsychiatry. 2015 Jul;1((2)):116–23. doi: 10.1159/000430916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Oguro-Ando A, Rosensweig C, Herman E, Nishimura Y, Werling D, Bill BR, et al. Increased CYFIP1 dosage alters cellular and dendritic morphology and dysregulates mTOR. Mol Psychiatry. 2014;20((9)):1069–78. doi: 10.1038/mp.2014.124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Trent S, Hall J, Connelly WM, Errington AC. Cyfip1 haploinsufficiency does not alter GABAA receptor δ-subunit expression and tonic inhibition in dentate gyrus PV+ interneurons and granule cells. eNeuro. 2019;6((3)):ENEURO.0364–18.2019. doi: 10.1523/ENEURO.0364-18.2019. Available from: http://www.ncbi.nlm.nih.gov/pubmed/31209152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Silva AI, Haddon JE, Ahmed Syed Y, Trent S, Lin TE, Patel Y, et al. Cyfip1 haploinsufficient rats show white matter changes, myelin thinning, abnormal oligodendrocytes and behavioural inflexibility. Nat Commun. 2019;10((1)):3455. doi: 10.1038/s41467-019-11119-7. Available from: https://www.ncbi.nlm.nih.gov/pubmed/31371763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Bachmann SO, Sledziowska M, Cross E, Kalbassi S, Waldron S, Chen F, et al. Behavioral training rescues motor deficits in Cyfip1 haploinsufficiency mouse model of autism spectrum disorders. Transl Psychiatry. 2019;9((1)):29. doi: 10.1038/s41398-018-0338-9. Available from: http://www.ncbi.nlm.nih.gov/pubmed/30664619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Fricano-Kugler C, Gordon A, Shin G, Gao K, Nguyen J, Berg J, et al. CYFIP1 overexpression increases fear response in mice but does not affect social or repetitive behavioral phenotypes. Mol Autism. 2019;10((1)):25. doi: 10.1186/s13229-019-0278-0. Available from: http://www.ncbi.nlm.nih.gov/pubmed/31198525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Stone JL, O'Donovan MC, Gurling H, Kirov GK, Blackwood DHR, Corvin A, et al. Rare chromosomal deletions and duplications increase risk of schizophrenia. Nature. 2008;455((7210)):237–41. doi: 10.1038/nature07239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Stefansson H, Rujescu D, Cichon S, Pietiläinen OP, Ingason A, Steinberg S, et al. Large recurrent microdeletions associated with schizophrenia. Nature. 2008;455((7210)):232–6. doi: 10.1038/nature07229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Kirov G, Grozeva D, Norton N, Ivanov D, Mantripragada KK, Holmans P, et al. Support for the involvement of large copy number variants in the pathogenesis of schizophrenia. Hum Mol Genet. 2009;18((8)):1497–503. doi: 10.1093/hmg/ddp043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Tam GW, van de Lagemaat LN, Redon R, Strathdee KE, Croning MD, Malloy MP, et al. Confirmed rare copy number variants implicate novel genes in schizophrenia. Biochem Soc Trans. 2010;38((2)):445–51. doi: 10.1042/BST0380445. Available from: http://www.ncbi.nlm.nih.gov/pubmed/20298200. [DOI] [PubMed] [Google Scholar]
- 209.Zhao Q, Li T, Zhao X, Huang K, Wang T, Li Z, et al. Rare CNVs and tag SNPs at 15q11.2 are associated with schizophrenia in the Han Chinese population. Schizophr Bull. 2013;39((3)):712–9. doi: 10.1093/schbul/sbr197. Available from: http://www.ncbi.nlm.nih.gov/pubmed/22317777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Grozeva D, Conrad DF, Barnes CP, Hurles M, Owen MJ, O'Donovan MC, et al. Independent estimation of the frequency of rare CNVs in the UK population confirms their role in schizophrenia. Schizophr Res. 2012 Mar;135((1–3)):1–7. doi: 10.1016/j.schres.2011.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.de Kovel CG, Trucks H, Helbig I, Mefford HC, Baker C, Leu C, et al. Recurrent microdeletions at 15q11.2 and 16p13.11 predispose to idiopathic generalized epilepsies. Brain. 2010 Jan;133((Pt 1)):23–32. doi: 10.1093/brain/awp262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Cooper GM, Coe BP, Girirajan S, Rosenfeld J, Vu TH, Baker C, et al. A copy number variation morbidity map of developmental delay. Nat Genet. 2011;43((9)):838–46. doi: 10.1038/ng.909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Chaste P, Sanders SJ, Mohan KN, Klei L, Song Y, Murtha MT, et al. Modest impact on risk for autism spectrum disorder of rare copy number variants at 15q11.2, specifically breakpoints 1 to 2. Autism Res. 2014 Jun;7((3)):355–62. doi: 10.1002/aur.1378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.von der Lippe C, Rustad C, Heimdal K, Rødningen OK. 15q11.2 microdeletion: seven new patients with delayed development and/or behavioural problems. Eur J Med Genet. 2011;54((3)):357–60. doi: 10.1016/j.ejmg.2010.12.008. Available from: http://www.ncbi.nlm.nih.gov/pubmed/21187176. [DOI] [PubMed] [Google Scholar]
- 215.Gudmundsson OO, Walters GB, Ingason A, Johansson S, Zayats T, Athanasiu L, et al. Attention-deficit hyperactivity disorder shares copy number variant risk with schizophrenia and autism spectrum disorder. Transl Psychiatry. 2019 Oct;9((1)):258. doi: 10.1038/s41398-019-0599-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Zhang X, Abdellaoui A, Rucker J, de Jong S, Potash JB, Weissman MM, et al. Genome-wide burden of rare short deletions is enriched in major depressive disorder in four cohorts. Biol Psychiatry. 2019 Jun;85((12)):1065–73. doi: 10.1016/j.biopsych.2019.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.van der Zwaag B, Staal WG, Hochstenbach R, Poot M, Spierenburg HA, de Jonge MV, et al. A co-segregating microduplication of chromosome 15q11.2 pinpoints two risk genes for autism spectrum disorder. Am J Med Genet B Neuropsychiatr Genet. 2010;153((4)):960–6. doi: 10.1002/ajmg.b.31055. Available from: http://www.ncbi.nlm.nih.gov/pubmed/20029941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Wang J, Tao Y, Song F, Sun Y, Ott J, Saffen D. Common regulatory variants of CYFIP1 contribute to susceptibility for autism spectrum disorder (ASD) and classical autism. Ann Hum Genet. 2015 Sep;79((5)):329–40. doi: 10.1111/ahg.12121. [DOI] [PubMed] [Google Scholar]
- 219.Waltes R, Duketis E, Knapp M, Anney RJ, Huguet G, Schlitt S, et al. Common variants in genes of the postsynaptic FMRP signalling pathway are risk factors for autism spectrum disorders. Hum Genet. 2014;133((6)):781–92. doi: 10.1007/s00439-013-1416-y. Available from http://www.ncbi.nlm.nih.gov/pubmed/24442360. [DOI] [PubMed] [Google Scholar]
- 220.Hirayama-Kurogi M, Takizawa Y, Kunii Y, Matsumoto J, Wada A, Hino M, et al. Downregulation of GNA13-ERK network in prefrontal cortex of schizophrenia brain identified by combined focused and targeted quantitative proteomics. J Proteomics. 2017 Mar;158:31–42. doi: 10.1016/j.jprot.2017.02.009. [DOI] [PubMed] [Google Scholar]
- 221.Sledziowska M, Galloway J, Baudouin S. Evidence for a contribution of the Nlgn3/Cyfip1/Fmr1 pathway in the pathophysiology of autism spectrum disorders. Neuroscience. 2019 Nov doi: 10.1016/j.neuroscience.2019.10.011.0. In press. [DOI] [PubMed] [Google Scholar]