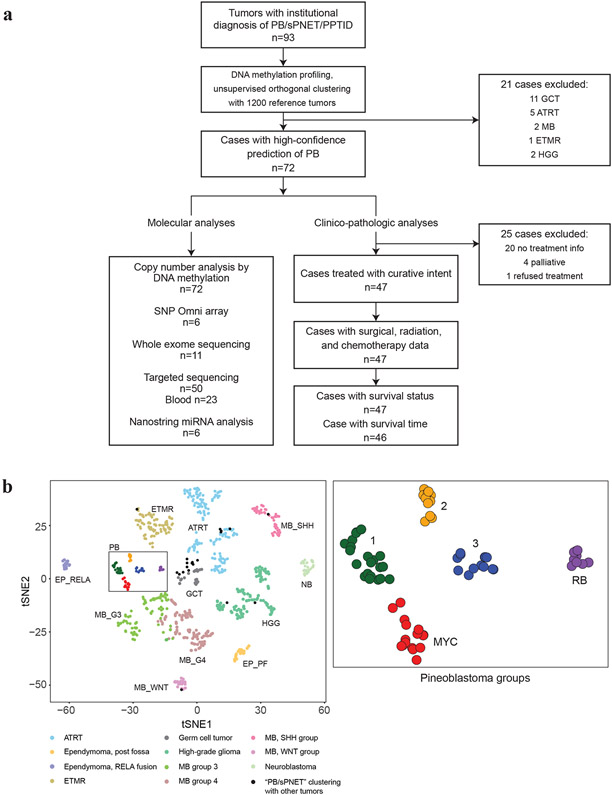
Figure 1. PB comprise five molecular sub-groups.
a. Flow diagram of analyses performed: 93 primary tumors with institutional diagnosis of pineoblastoma (PB) or supratentorial PNET (sPNET) were analysed using global methylation profiling and compared against a reference cohort of 1200 pediatric brain tumors to identify and exclude samples that segregated with other brain tumors. A cluster of robust, molecularly confirmed 72 PBs were further characterized using methylation and SNP arrays for copy number alterations, mutational analyses using WES and targeted sequencing, and Nanostring analyses for miRNA expression. Clinical, treatment, and molecular sub-group data available for 46 PB patients treated with curative intent were integrated for clinic-pathologic analyses.
b. t-Distributed stochastic neighbour embedding (tSNE) plots of DNA methylation clustering patterns of 93 presumed PB samples relative to 951/1200 representative pediatric brain tumor entities demonstrate PB clusters separately from other tumor entities. Plots using the top 12,500 most varying methylation probes by standard deviation (SD) are shown. Tumors are shown as colored spheres which include atypical teratoid rhabdoid tumor (ATRT), ependymoma posterior-fossa (EP_PF) or supratentorial, RELA-fusion (EP_RELA), embryonal tumor multiple rosettes (ETMR), germ cell tumor (GCT), high-grade glioma (HGG), neuroblastoma (NB), medulloblastoma WNT (MB_WNT), SHH (MB_SHH), group 3 (MB_G3), and group 4 (MB_G4). Black spheres indicate tumors with an institutional diagnosis of PB that segregated with other known brain tumor entities are (n=21). A robust cluster of 72 PBs is boxed; blow-up image of PB cluster on right shows five molecular PB sub-groups designated as 1, 2, 3, RB, and MYC.