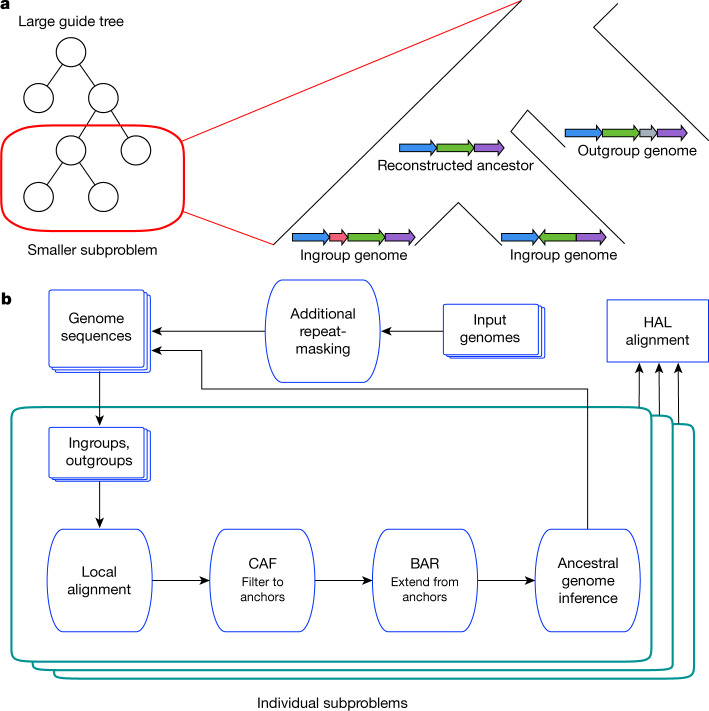
Fig. 1. The alignment process within Progressive Cactus.
a, A large alignment problem is split into many smaller subproblems using an input guide tree. Each subproblem compares a set of ingroup genomes (the children of the internal node to be reconstructed) against each other as well as a sample of outgroup genomes (non-descendants of the internal node in question). b, Flowchart represents the phases in which the overall alignment, as well as each subproblem alignment, proceeds through. The end result is a new genome assembly that represents the Progressive Cactus reconstruction of the ancestral genome, and an alignment between this ancestral genome and its children. After all subproblems have been completed, the parent–child alignments are combined to create the full reference-free alignment in the HAL27 format.