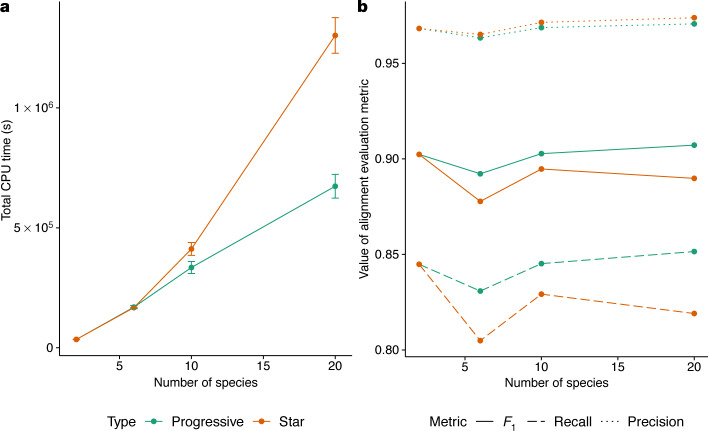
Fig. 2. Comparing alignments of varying numbers of simulated genomes using Progressive Cactus.
a, The progressive mode of Progressive Cactus is shown, versus the mode without progressive decomposition that is similar to that previously described6 (‘star’). The average total runtime of the two alignment methods across three runs is shown. Data are mean and s.d. The runtime is identical when aligning two genomes as the alignment problem is not further decomposed, but the linear scaling of the progressive mode means it is much faster with large numbers of genomes than the quadratic scaling required without progressive alignment. b, The precision, recall and F1 score (harmonic mean of precision and recall) of aligned pairs for each alignment compared with pairs from the true alignment produced by the simulation.