Fig. 2. Metadynamic metainference simulations characterize the dynamic binding and show how 10074-G5 promotes Aβ42 conformations with less hydrophobicity.
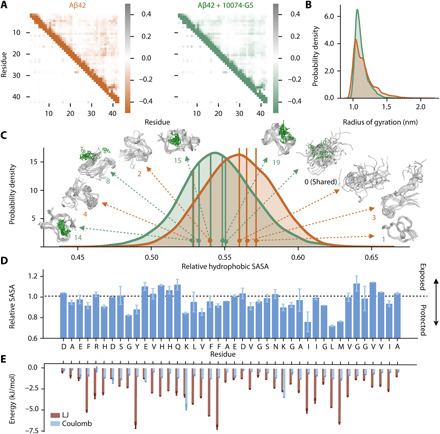
(A) Metadynamics metainference simulations demonstrate that inter-residue contact maps for Lennard-Jones (LJ) (top right) and Coulomb (top left) potentials for the unbound (orange) and the bound (green) structural ensembles of Aβ42 with 10074-G5 are highly similar. (B) Radii of gyration for the unbound and bound structural ensembles are also highly similar (shown using kernel density estimates of 35,000 points each sampled based on metadynamics weights using a Gaussian kernel). (C) Relative hydrophobic solvent accessible surface area (SASA) of Aβ42 (total hydrophobic area over total surface area) of the bound and unbound ensembles, showing that 10074-G5 decreases the relative exposed hydrophobicity of Aβ42. The holo ensemble was calculated only on the protein surface but accounts for the presence of the compound. Data are shown using kernel density estimates as described in (B). Some of the representative structures from these distributions are shown. Numbers indicate cluster IDs shown in Fig. 3. (D) Ratio (bound/unbound) of the ensemble-averaged, total SASA per residue showing regions of Aβ42 that become more exposed or protected in the presence of 10074-G5. SASAs of the bound ensemble were calculated on the protein in the presence of 10074-G5. (E) Ensemble-averaged, residue-specific LJ and Coulomb interaction energies show that 10074-G5 has strong interactions with aromatic and charged residues. Error bars represent SDs between first and second halves of the analyzed trajectories in (D) and (E).
