Fig. 3. The motif binds to Rubisco.
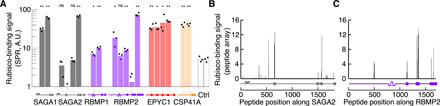
(A) Peptides containing motifs from the indicated proteins were synthesized, and their binding to Rubisco was measured by surface plasmon resonance (SPR). A.U., arbitrary units. Two peptides not containing the motif were included as controls (Ctrl). Each dot shows the binding response of an independent replica. Negative values (when the experimental binding signal was lower than that of the reference cell) are not plotted. The positions of the predicted motifs are indicated below the graph (not to scale). Significance levels of increased binding relative to control peptides were determined using Welch’s t test. *P < 0.05 and **P < 0.01. (B and C) Arrays of 18–amino acid peptides tiling across the sequences of SAGA2 (B) and RBMP2 (C) were synthesized and probed with Rubisco. The binding signal in (B) and (C) is normalized to a control EPYC1 peptide known to bind to Rubisco (one unit of binding) (21). The positions of motifs are indicated below each graph (to scale).
